Large-scale genetic typing method based on SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology
A genotyping method and large-scale technology, applied in the field of large-scale sample genotyping, which can solve the problems of low accuracy and low throughput
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Example 1 The large-scale sample genotyping method based on sequencing and SLAFseq technology was applied to the construction of the genetic map of 224 carp F1 populations
[0050] Carp genetic map construction, including the following steps:
[0051] 1. Selection of carp colony (purchased from Heilongjiang Fisheries Research Institute): the colony is F1 colony with 224 strains, 2 parent strains, and 50,000 tags developed.
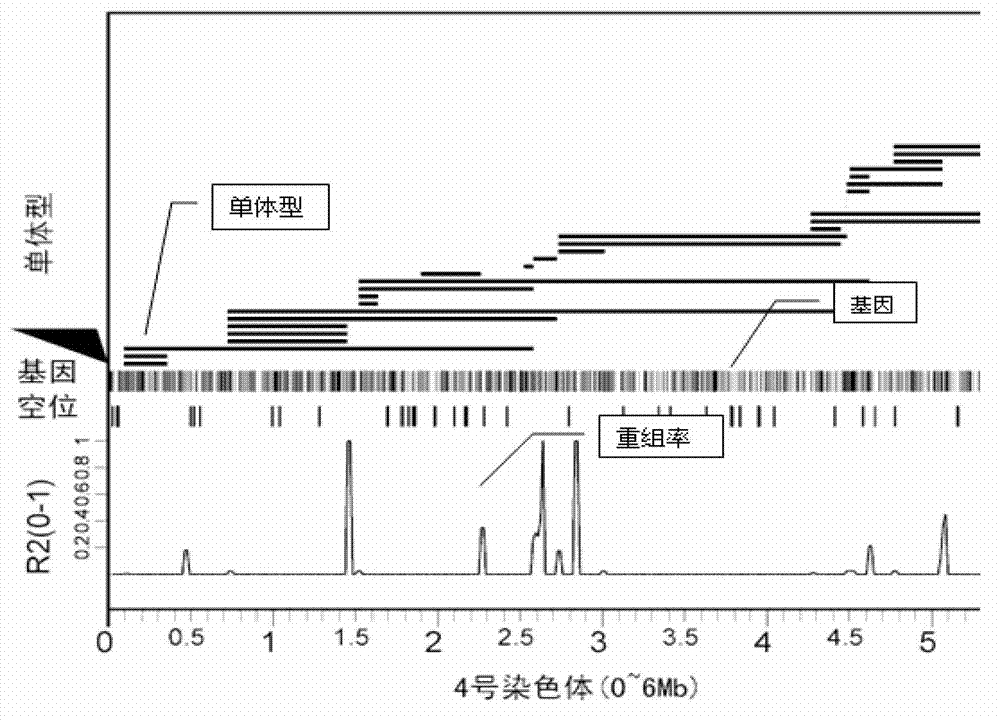
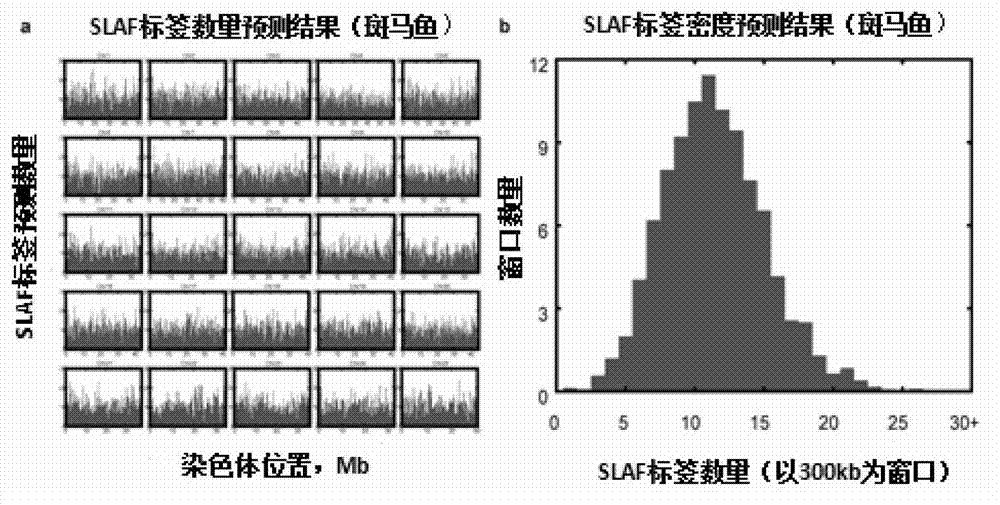
[0052] 2. As shown in Table 1, the bioinformatics method is used to predict the genome sequence of a similar species of zebrafish, the genome version number is Zv9 published by Ensembl, and the evaluation results of the parameters of the selected optimal solution AluI+MseI, The results showed that the number of enzyme-digested fragments with a specific length of 330-380 was 64,918, which met the needs of tag development, and the ratio of low-copy tags was 94.97%, effectively avoiding highly repetitive sequences; Figure 1A Shown is the evaluation ma...
Embodiment 2
[0077] Example 2 The large-scale sample genotyping method based on sequencing and SLAFseq technology is applied to the construction of haplotype maps and trait association analysis of 512 natural soybean populations
[0078] The soybean haplotype map construction and trait association analysis include the following steps:
[0079]1. Selection of soybean populations: There are 512 germplasm resources of soybean natural populations, and 50,000 SLAF tags are designed.
[0080] 2. If Figure 7 Shown is the evaluation diagram of the effect of avoiding highly repetitive regions of the optimal scheme HaeIII+MseI determined by the bioinformatics simulation of the soybean genome combined with the preliminary experiment. The results show that the digestion fragments of this scheme are evenly distributed on the genome and Effectively reduce the proportion of repetitive sequences in the genome; as shown in Table 6, the genome simplification effect evaluation statistics for this program s...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com