Patents
Literature
73 results about "Genomewide association" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
Large-scale genetic typing method based on SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology
ActiveCN103088120AGuarantee label qualityQuality assuranceMicrobiological testing/measurementGenotyping by sequencingGenome
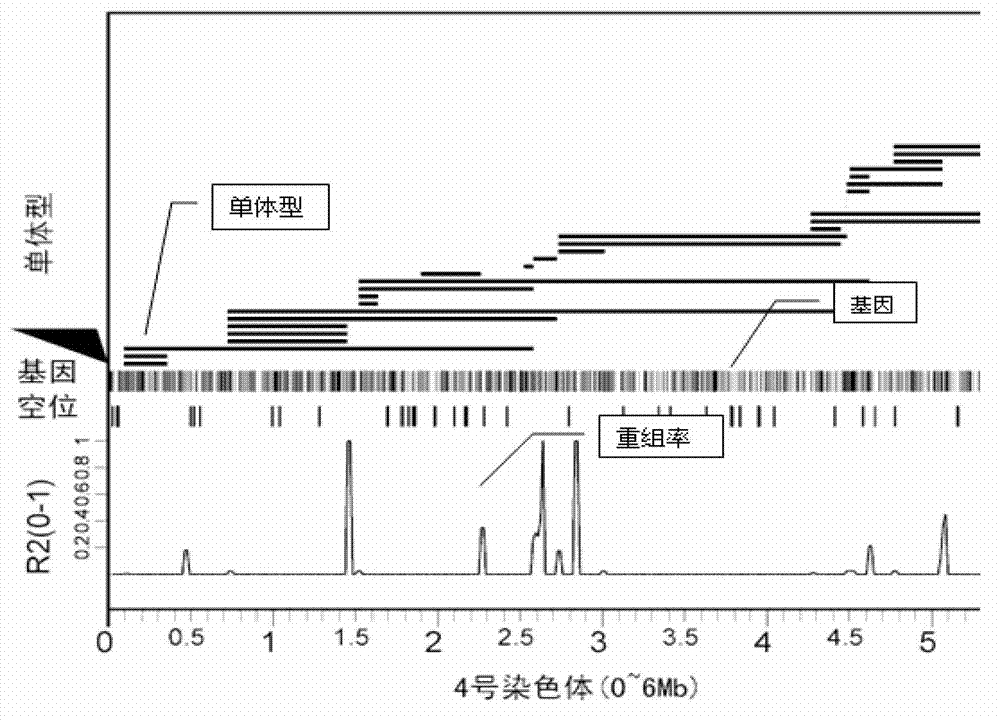
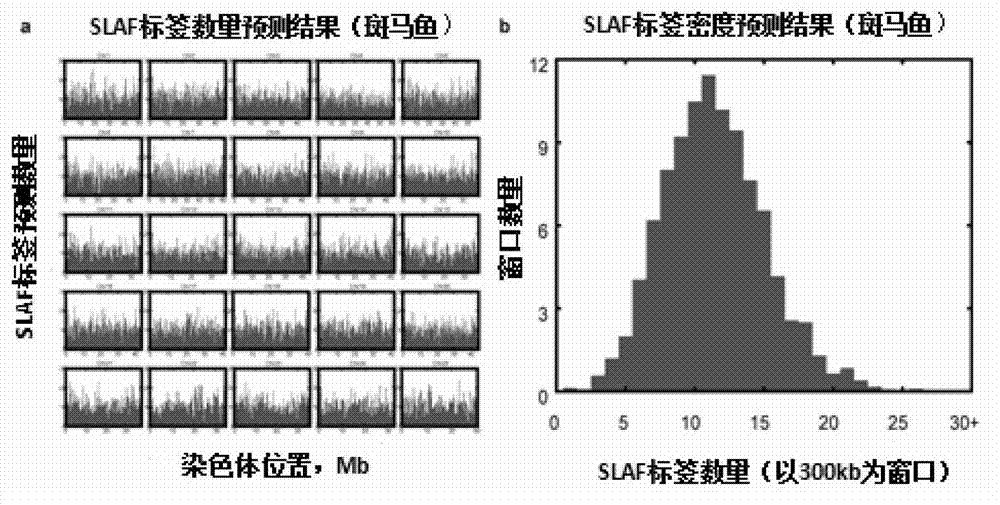
The invention provides a method for carrying out large-scale gene typing based on an n SLAF-seq (Specific-Locus Amplified Fragment Sequencing) technology. Complexity of the genome is reduced by utilizing the SLAF-seq technology, and genetic typing is carried out on large-scale products. High-throughput sequencing is carried out on the genome, marker-developing, genetic map drawing and whole genome association analyzing are carried out on the samples by utilizing the technology. Compared with the conventional method, the large-scale genetic typing method disclosed by the invention has the advantages that the throughput is greatly improved, and the cost is greatly reduced. The method is mainly applied to marker-developing, genetic map drawing and whole genome association analyzing.
Owner:BIOMARKER TECH
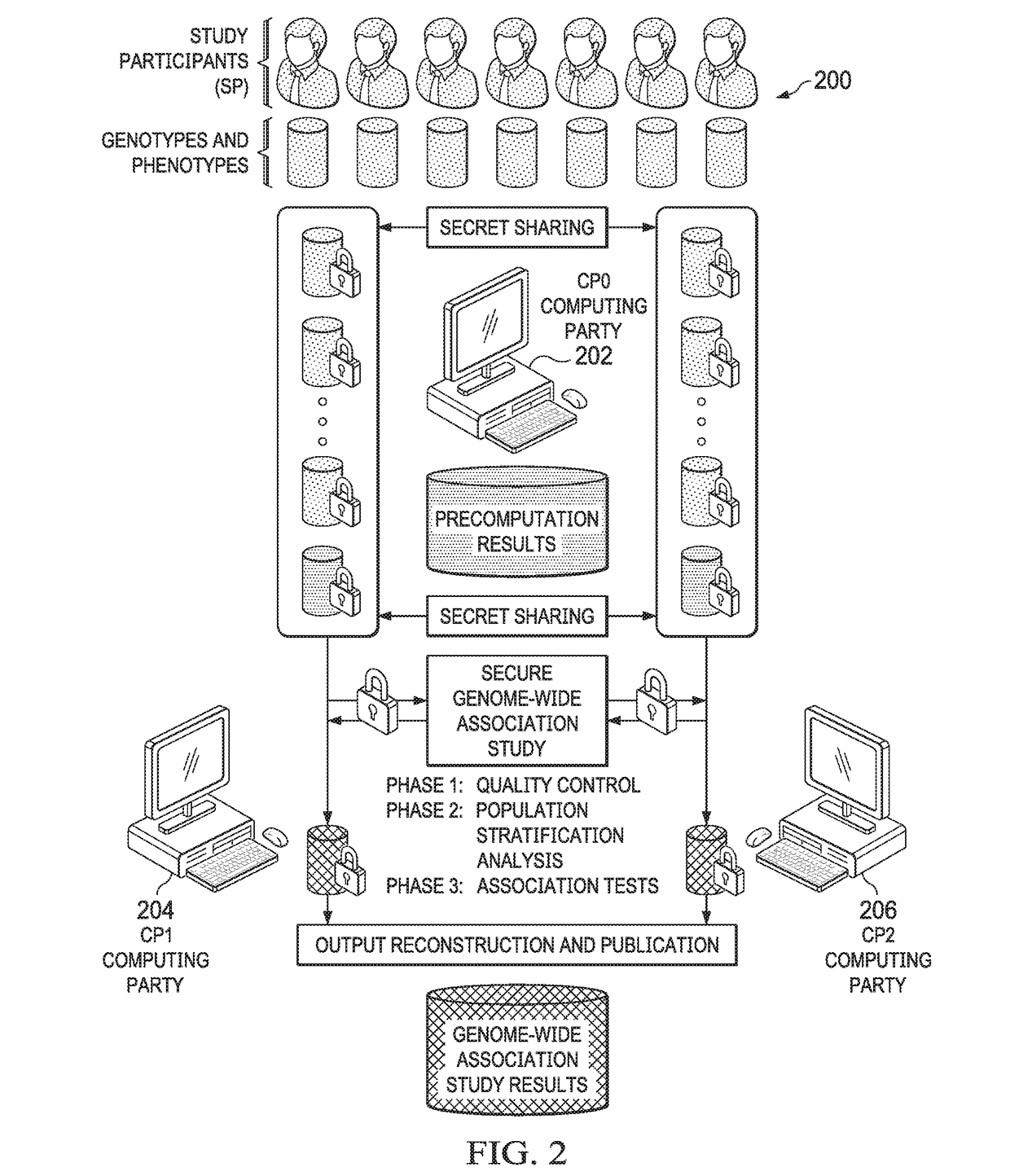
Secure genome crowdsourcing for large-scale association studies
ActiveUS20180373834A1Secure crowdsourcingPrivacy protectionProteomicsGenomicsData setIndividual study
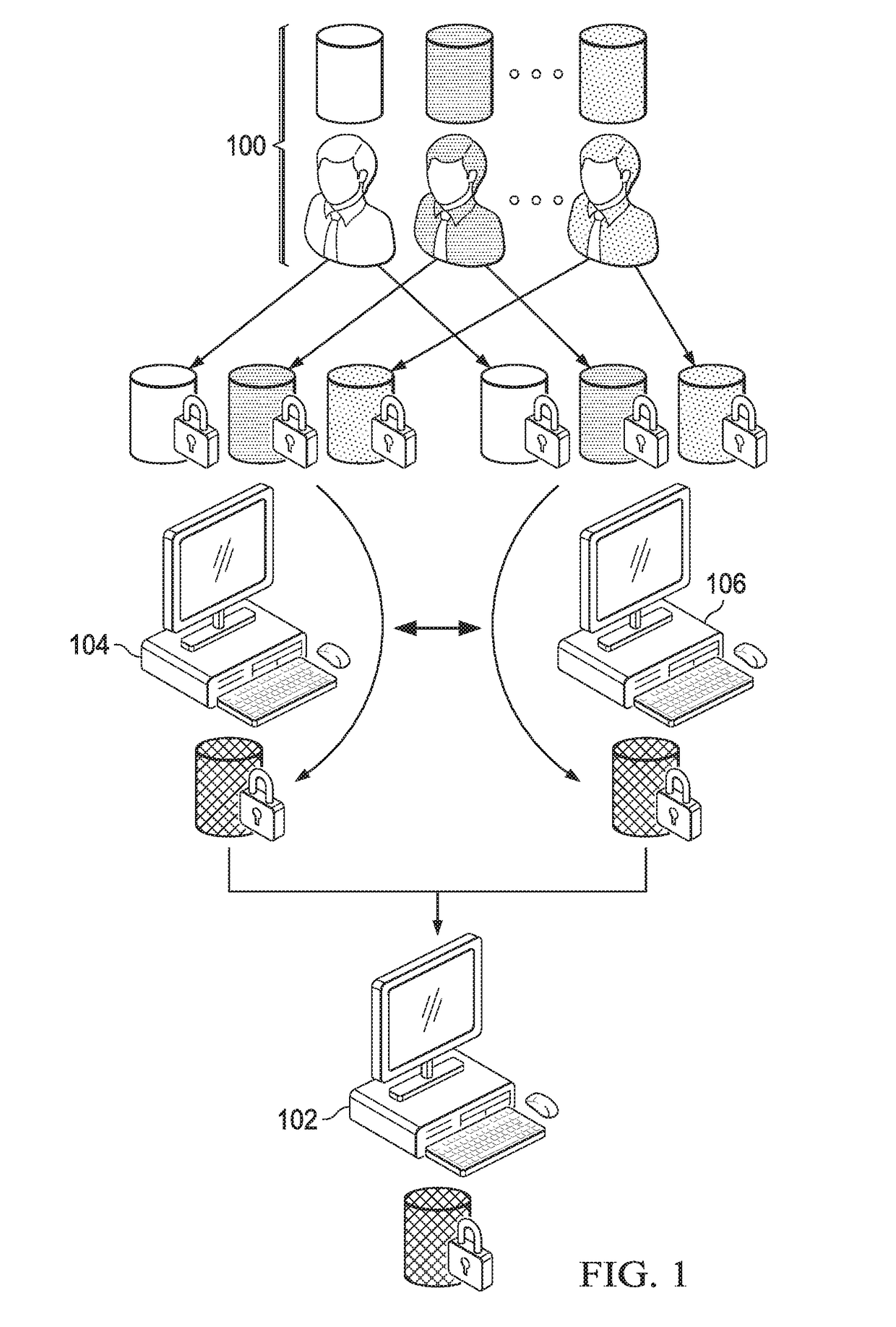
Computationally-efficient techniques facilitate secure crowdsourcing of genomic and phenotypic data, e.g., for large-scale association studies. In one embodiment, a method begins by receiving, via a secret sharing protocol, genomic and phenotypic data of individual study participants. Another data set, comprising results of pre-computation over random number data, e.g., mutually independent and uniformly-distributed random numbers and results of calculations over those random numbers, is also received via secret sharing. A secure computation then is executed against the secretly-shared genomic and phenotypic data, using the secretly-shared results of the pre-computation over random number data, to generate a set of genome-wide association study (GWAS) statistics. For increased computational efficiency, at least a part of the computation is executed over dimensionality-reduced genomic data. The resulting GWAS statistics are then used to identify genetic variants that are statistically-correlated with a phenotype of interest.
Owner:CHO HYUNGHOON +2
Genome-Wide Association Study Identifying Determinants Of Facial Characteristics For Facial Image Generation
InactiveUS20130039548A1Promote generationBiometric pattern for forensic purposeImage enhancementA-DNAOrganism
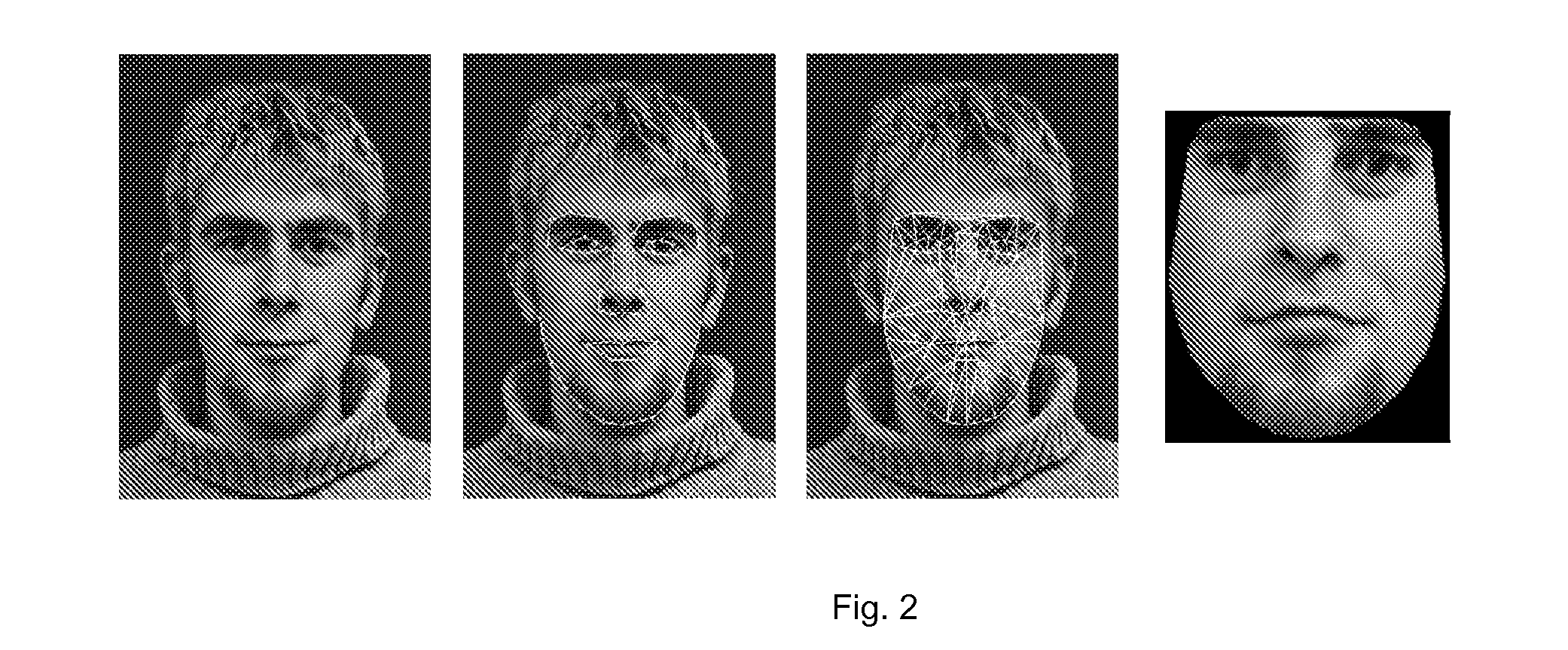
The present invention relates to a method for the generation of a facial composite from the genetic profile of a DNA-donor. The method comprises the steps of a) subjecting a biological sample to genotyping thereby generating a profile of genetic markers associated to numerical facial descriptors (NFD) for said sample, b) reverse engineer a NFD from the profile of the associated genetic variants and constructing a facial composite from the reverse engineered numerical facial descriptors (NFDs). The present invention also relates to a method for identifying genetic markers and / or combinations of genetic markers that are predictive of the facial characteristics, (predictive facial markers) of a person, said method comprising the steps of: a) capturing images of a group of individual faces; b) performing image analysis on facial images of said group of individual faces thereby extracting phenotypical descriptors of the faces; c) obtaining data on genetic variation from said group of individual and d) performing a genome-wide association study (GWAS) to identify said predictive facial markers.
Owner:DANMARKS TEKNISKE UNIV
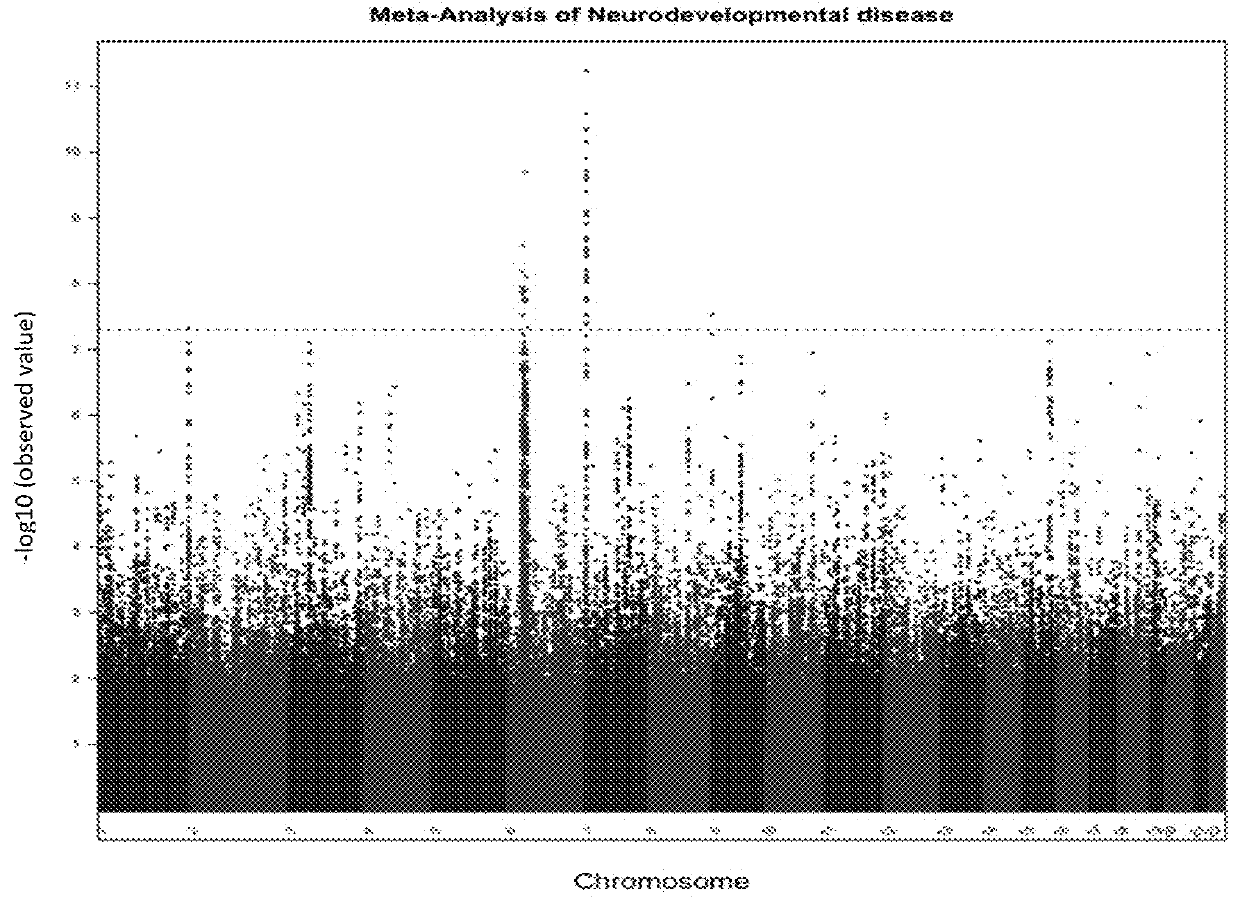
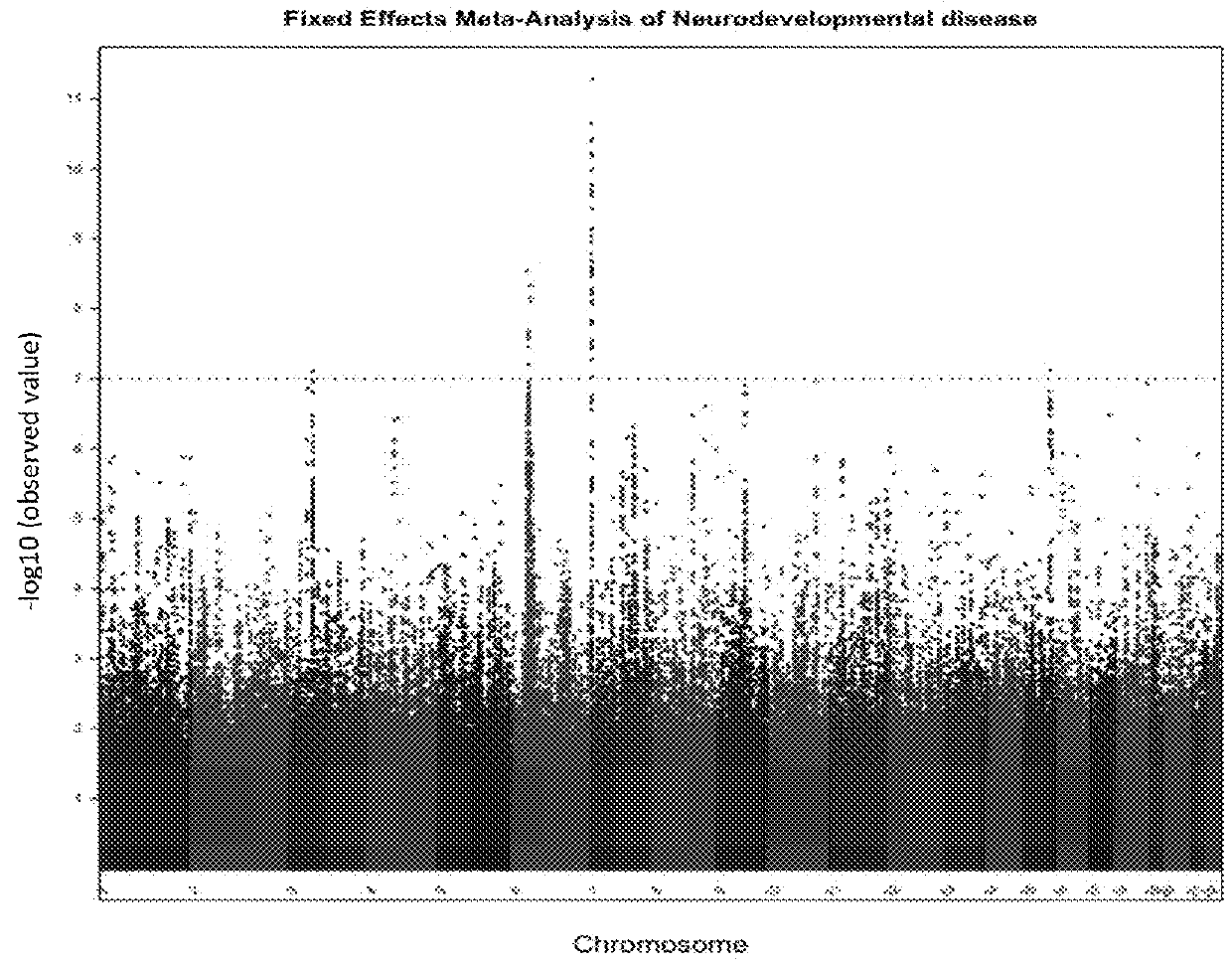
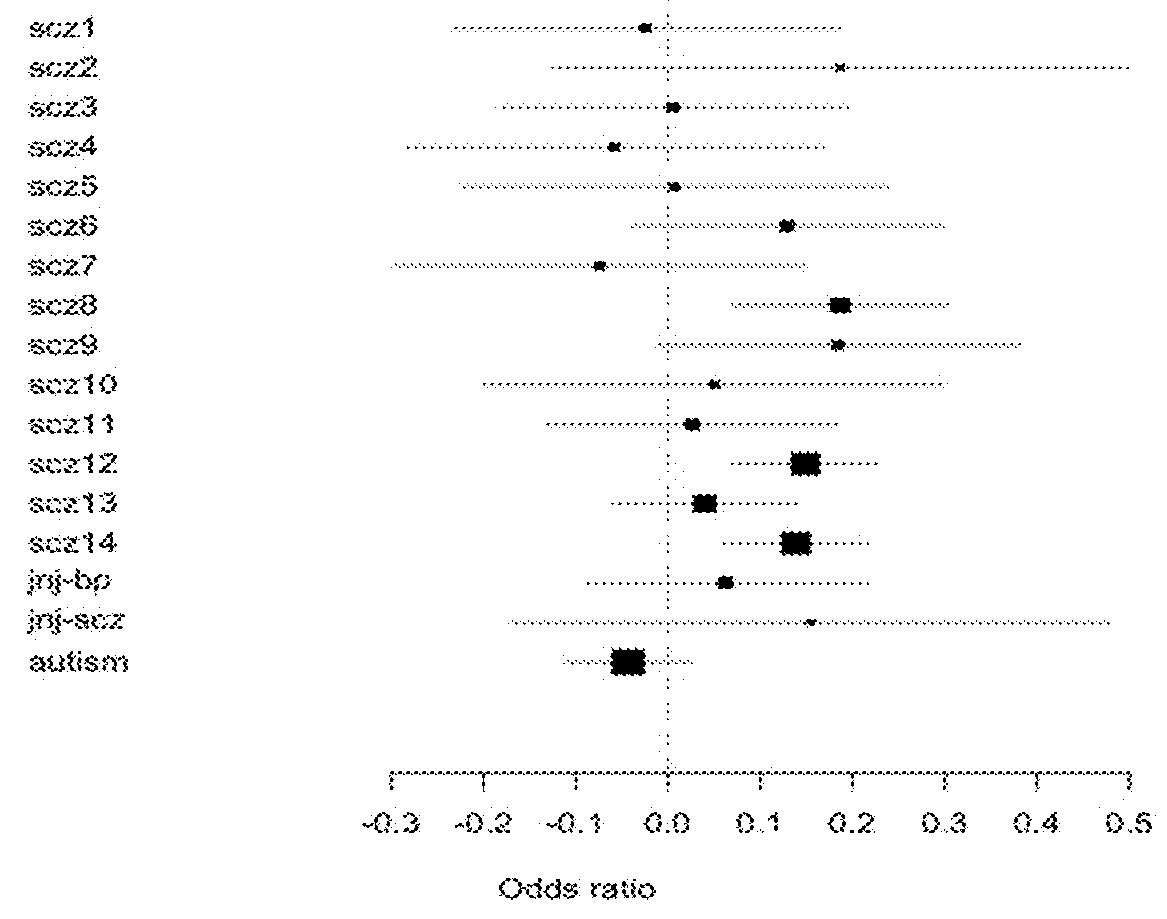
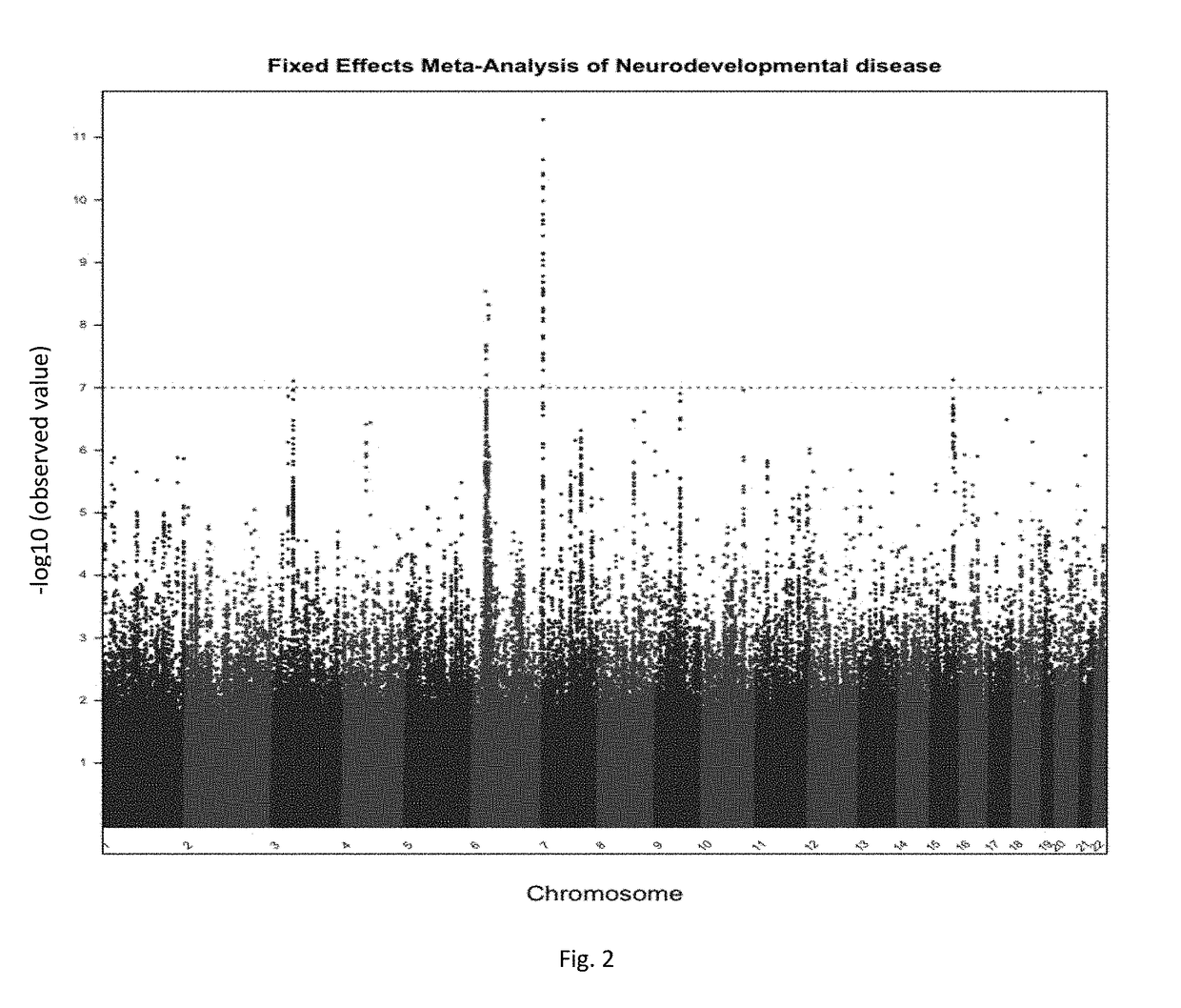
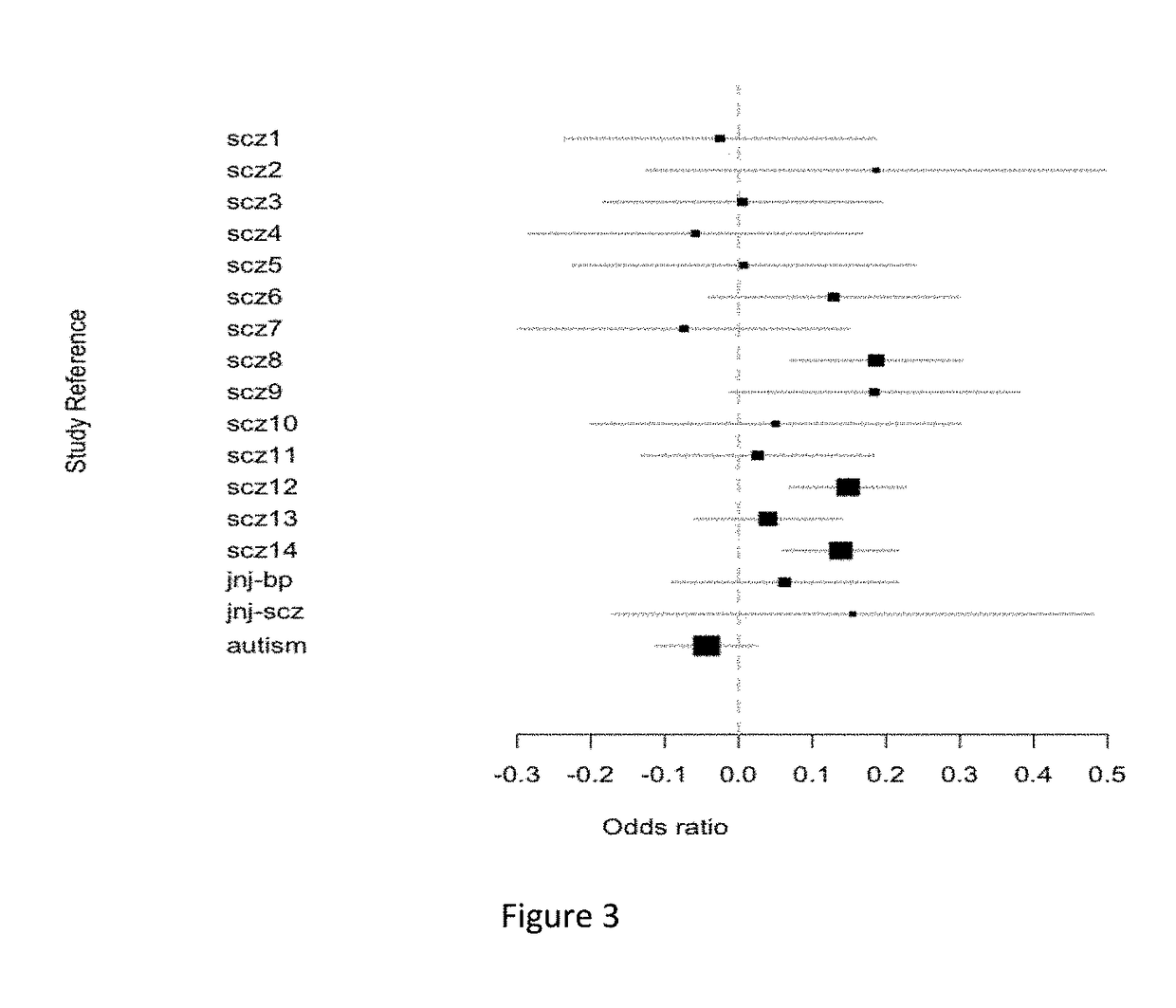
Schizophrenia-associated genetic loci identified in genome wide association studies and methods of use thereof
Compositions and methods for the identification of agents useful for the treatment of neurological disorders, including schizophrenia, are provided.
Owner:THE CHILDRENS HOSPITAL OF PHILADELPHIA
Genemap of the human genes associated with crohn's disease
InactiveUS20100081129A1Genetic material ingredientsMicrobiological testing/measurementLinkage Disequilibrium MappingPharmacogenomic Study
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to Crohn's disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
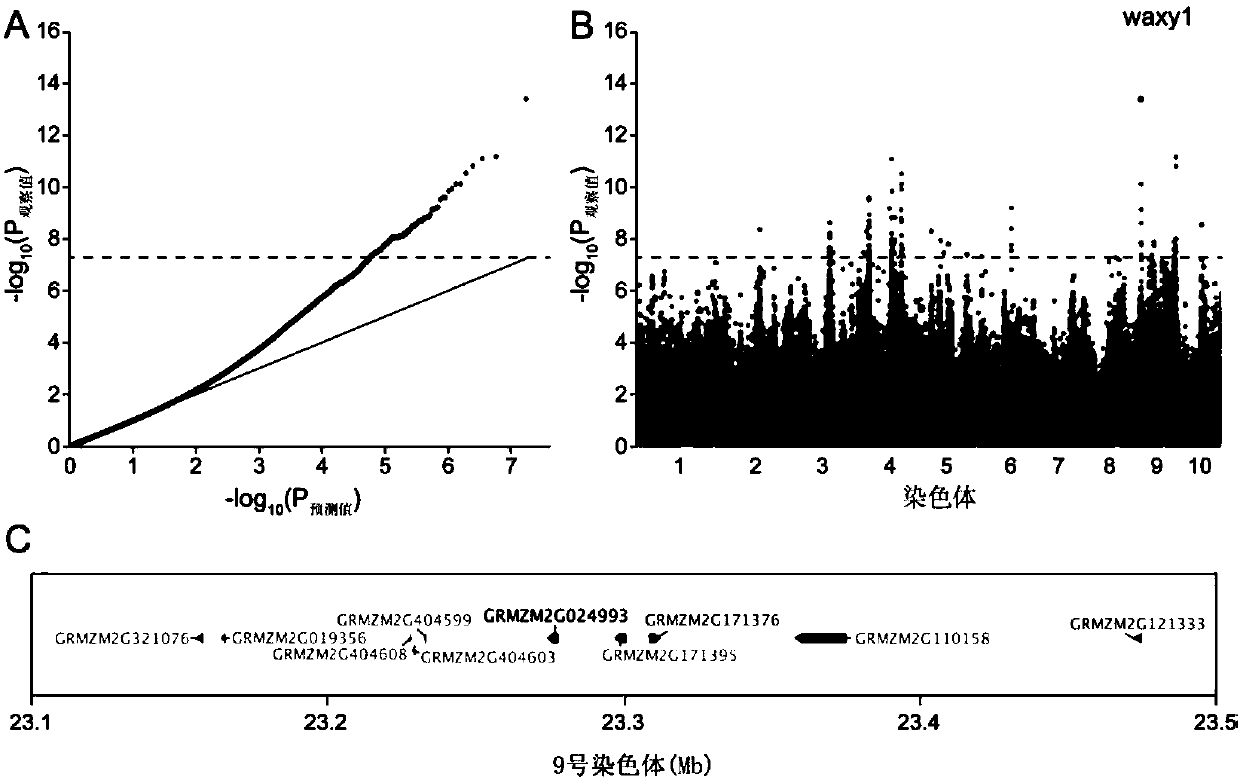
Method for developing waxy1-gene internal molecular markers on basis of association analysis and KASP
ActiveCN107619873AQuick digRapid phenotypeMicrobiological testing/measurementDNA/RNA fragmentationAmylaseCandidate Gene Association Study
The invention discloses a method for developing waxy1-gene internal molecule markers on the basis of association analysis and KASP. One indel and two snap markers located in waxy1 gene and remarkablyassociated with content of amylase are found out mainly through genome-wide association study and candidate gene association analysis techniques; the detected indel is marked as a major mutant type ofglutinous maize in China; glutinous maize materials can be identified through agarose gel by the markers developed on the basis of the indel, and convenience and rapidness is achieved as compared with that of original polyacrylamide gel identification; meanwhile, paired linkage disequilibrium test of 59 parts of waxy1-gene sequence regions of maize hybrid line materials is performed, close linkage of the indel marker and the two snp markers is tested, primers of the two snps are developed on the basis of the KASP technology, the glutinous maize materials can be identified rapidly, efficientlyand accurately, and molecular breeding of glutinous maize is facilitated.
Owner:SHANGHAI JIAO TONG UNIV
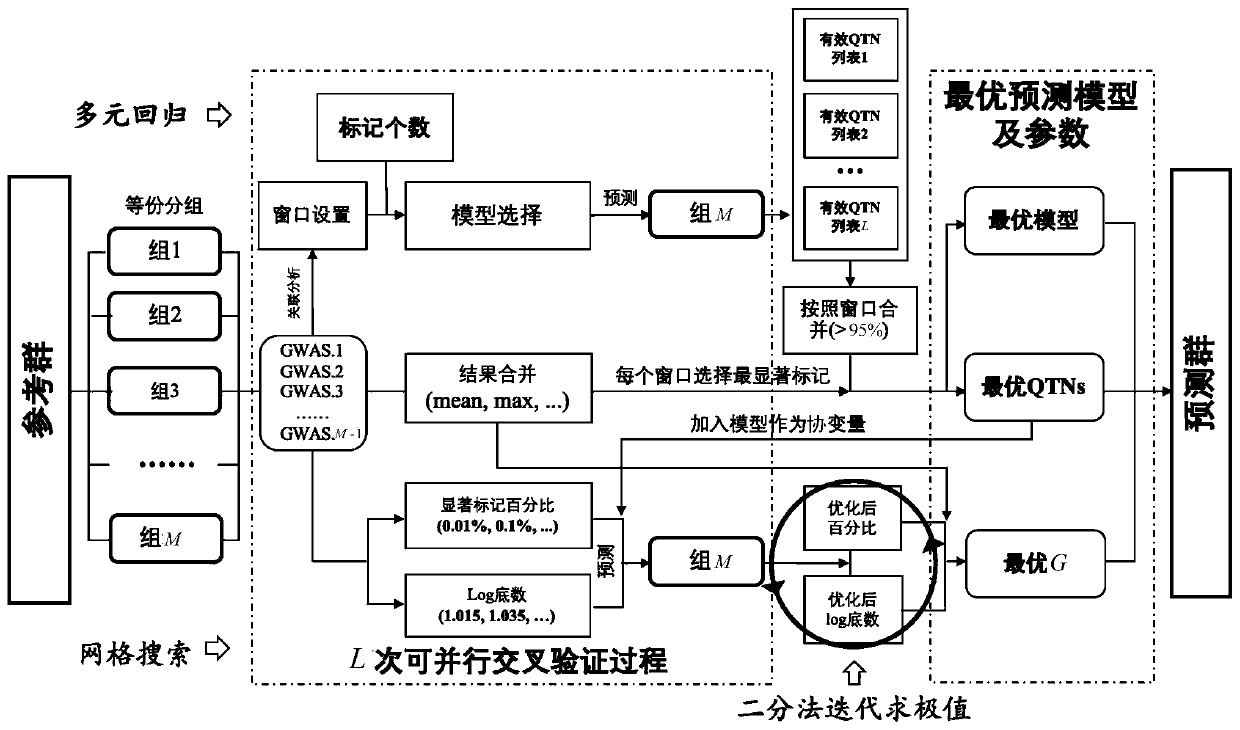
Efficient high-accuracy whole-genome selection method capable of performing parallel operation
ActiveCN110610744AImprove accuracyImprove computing efficiencyProteomicsGenomicsWhole Genome Association AnalysisAlgorithm
The invention relates to the technical field of animal and plant breeding and human disease prediction, and provides an efficient high-accuracy whole-genome selection method capable of performing parallel operation. The method comprises the following steps: firstly, reading an original genotype file and a phenotype file, constructing a new genotype file and a new phenotype file, and calculating agenetic relationship matrix of all individuals; then, extracting all individuals in the new phenotypic file as a reference group, and extracting all individuals without phenotypic data in the originalgenotypic file as a prediction group; carrying out whole genome association analysis by utilizing the reference group data, and extracting result characteristics of the whole genome association analysis; constructing a model library with specific characters, sequentially optimizing an optimal fixed effect and an optimal random effect by adopting a cross validation strategy, and selecting an optimal prediction model from the model library; and finally, calculating genome estimated breeding values of the prediction group by utilizing the optimal prediction model. The method can quickly, accurately and stably predict individual genome breeding values, and thus the accuracy and efficiency of whole genome selection are improved.
Owner:武汉影子基因科技有限公司
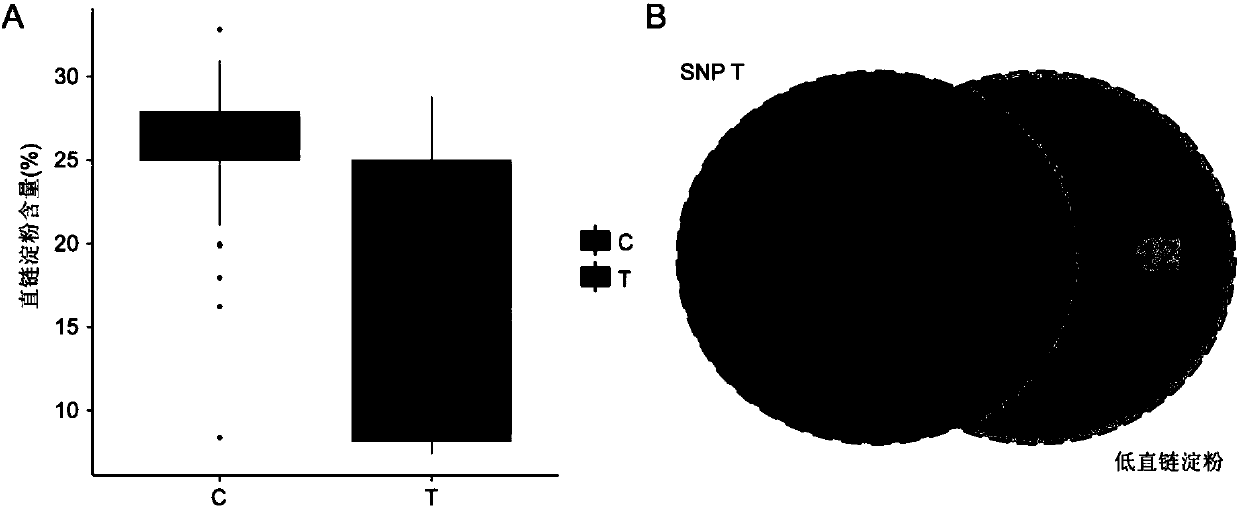
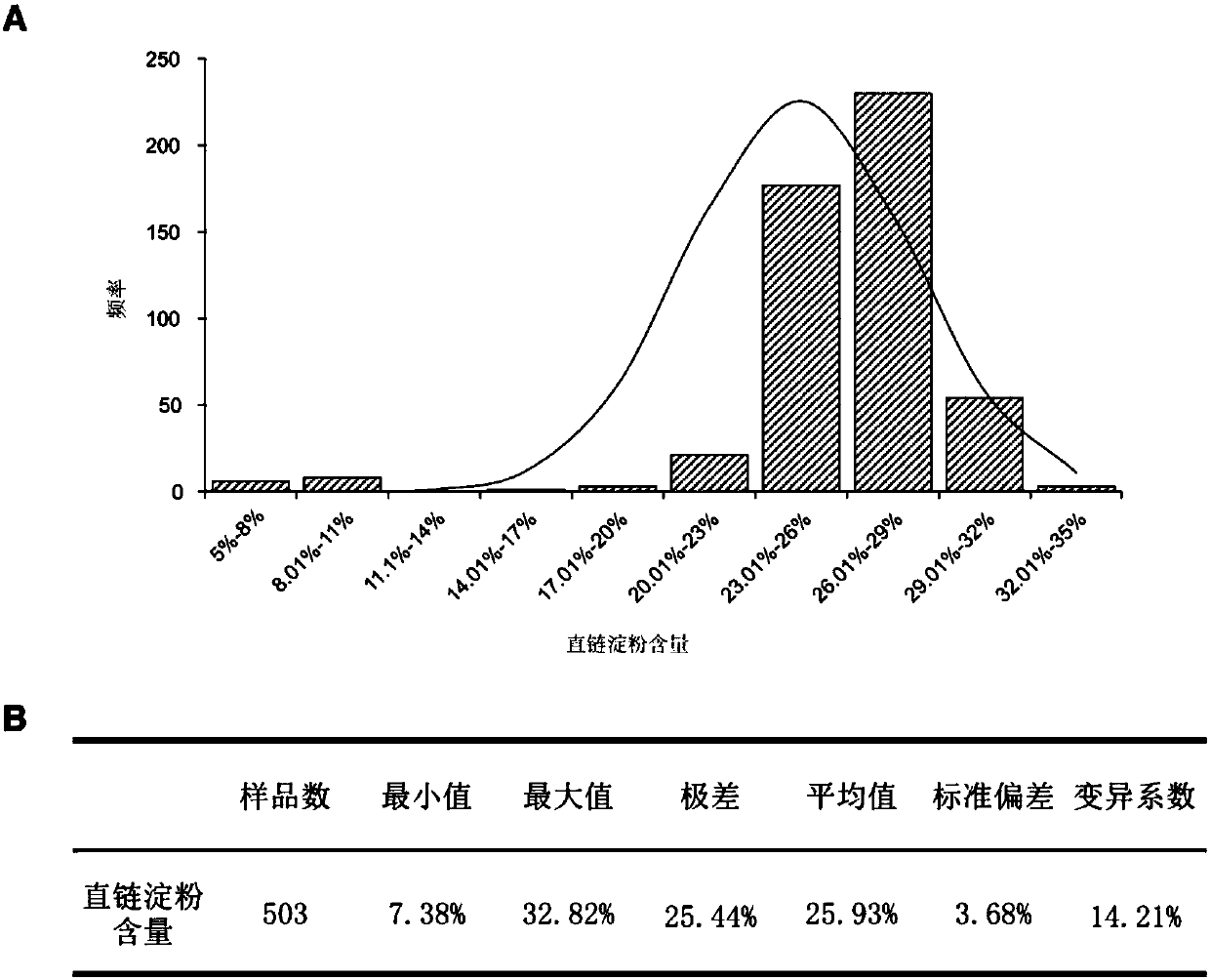
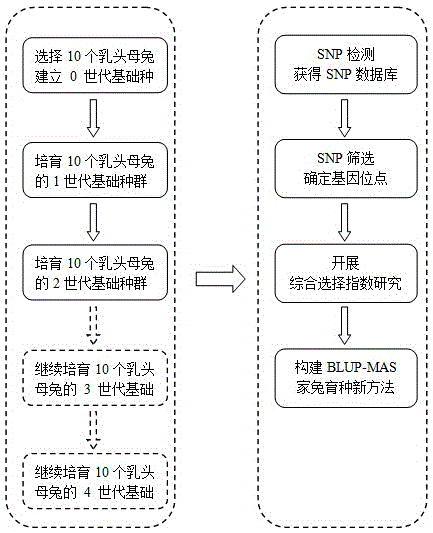
Method for breeding high-reproductive-performance breeding rabbits through SNP assistant selection breeding technology
The invention belongs to the technical field of livestock breeding and relates to a method for breeding high-reproductive-performance breeding rabbits based on SNP. The method comprises the following steps that firstly, resource group establishing and character measuring are conducted, wherein 1, resource group establishing is conducted, and 2, character measuring is conducted; secondly, target SNP selecting and whole genome correlation analyzing are conducted, wherein 1, genome SNP finding and whole genome SNP chip customizing are conducted, 2, whole genome correlation analyzing and verifying are conducted, and 3, SNP gathering is conducted; thirdly, a BLUP method is conducted, wherein a marginal effect value is calculated based on the principles of economics, and the animal model BLUP method is used for studying rabbit breeding value comprehensive selecting indexes; fourthly, a new M-BLUP rabbit breeding method is conducted, wherein the genetic effect of significant SNP locuses is estimated, molecule marking and BLUP are combined, and the new M-BLUP rabbit breeding method is established; fifthly, the new M-BLUP rabbit breeding method is used for breeding the high-reproductive-performance rabbits. The SNP assistant selection breeding method is adopted, special strains can be bred, and a high-reproductive-performance meat rabbit matched system can be established.
Owner:JILIN KANGDA FOOD CO LTD +1
Colorectal cancer susceptibility diagnostic kit and application of SNP (single nucleotide polymorphism) in preparation of diagnostic kit
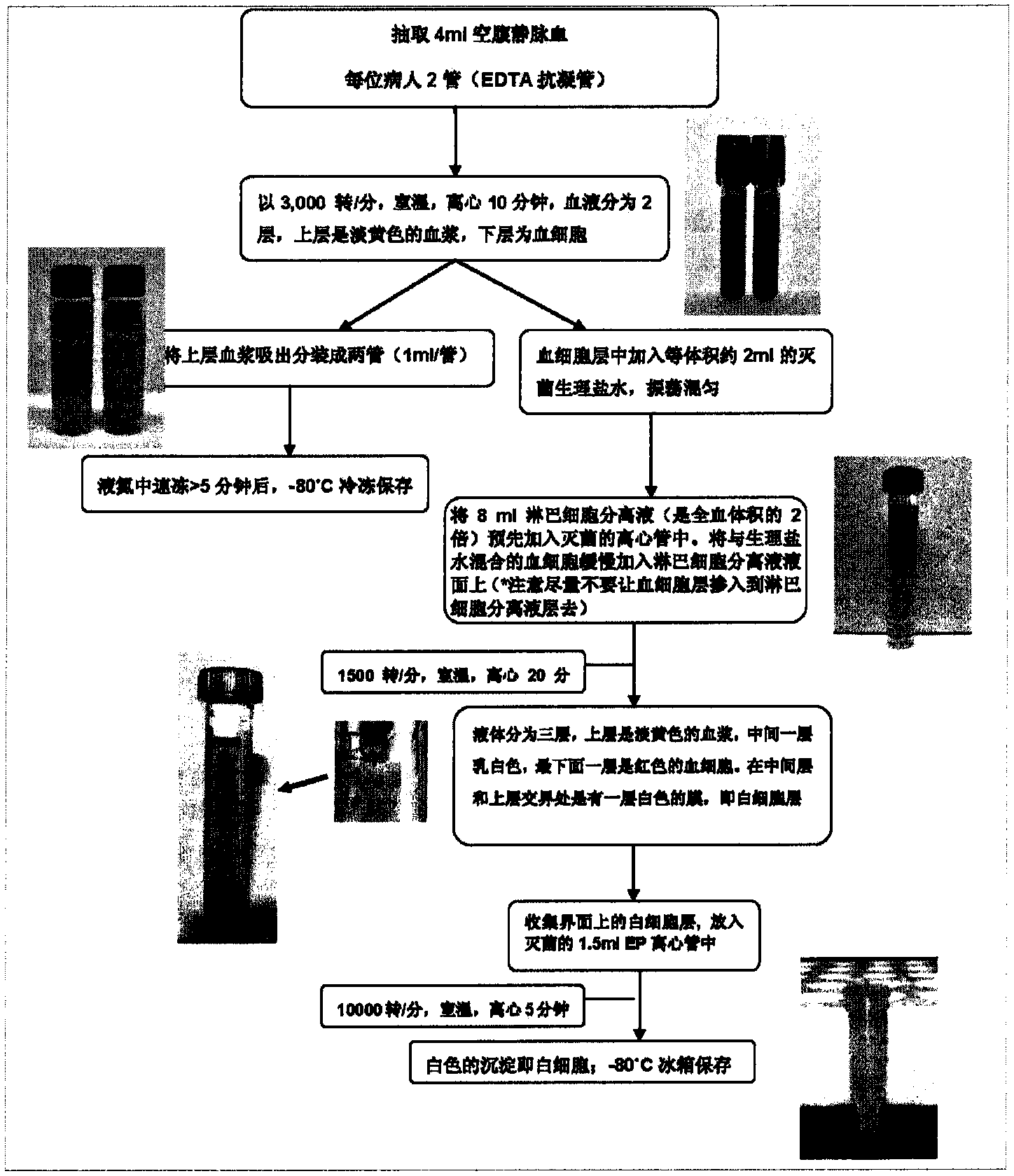
The invention discloses a colorectal cancer susceptibility diagnostic kit and an application of SNP (single nucleotide polymorphism) in preparation of the diagnostic kit. The SNP sites are rs12522693 and rs17836917. By means of genome-wide association study, the inventors prove remarkable correlation between an allele at the rs12522693 site G and an allele at the rs17836917 site A and colorectal cancer susceptibility through a screening stage, first stage verification and second stage verification. By means of detecting the genotypes of the rs12522693 site and / or the rs17836917 site, the colorectal cancer susceptibility of a testee can be predicted, and thus early prevention and treatment of the disease can be promoted. The colorectal cancer susceptibility diagnostic kit can be used for detecting the genotypes of the rs12522693 site and / or the rs17836917 site by using known technologies in the field so as to judge whether the testee has colorectal cancer susceptibility or not.
Owner:PEOPLES HOSPITAL PEKING UNIV
Method for discovering pharmacogenomic biomarkers
InactiveUS20140031242A1Quality improvementPredict likelihood of successSugar derivativesMicrobiological testing/measurementPharmacogenomicsDrugs response
The present invention relates to a method of discovering pharmacogenomic biomarkers that are correlated with varied individual responses (efficacy, adverse effect, and other end points) to therapeutic agents. The present invention provides a mean to utilize archived clinical samples to perform genome-wide association study in order to identify novel pharmacogenomic biomarkers. The newly discovered biomarkers can then be developed into companion diagnostic tests which can help to predict drug responses and apply drugs only to those who will be benefited, or exclude those who might have adverse effects, by the treatment.
Owner:DENOVO BIOPHARMA HANGZHOU LTD
Molecular marker influencing feed conversion ratio character of pig and application
ActiveCN105624155ALower conversion rateReduce manufacturing costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFeed conversion ratio
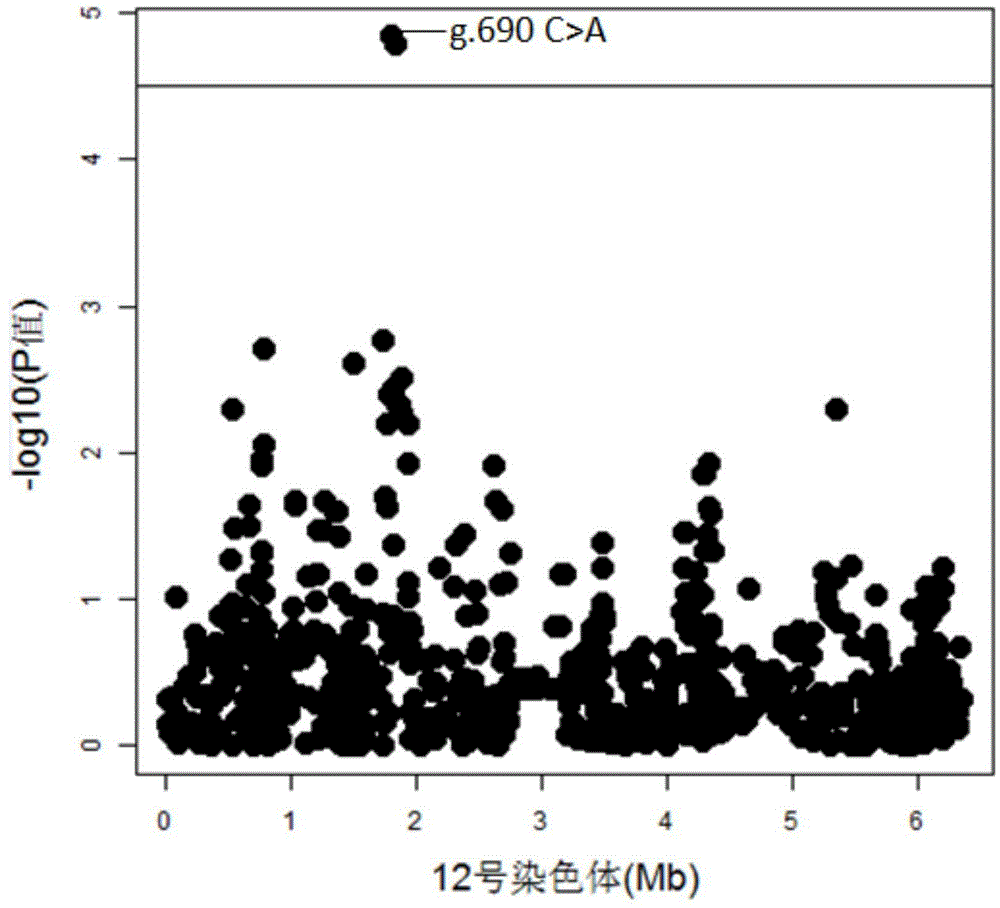
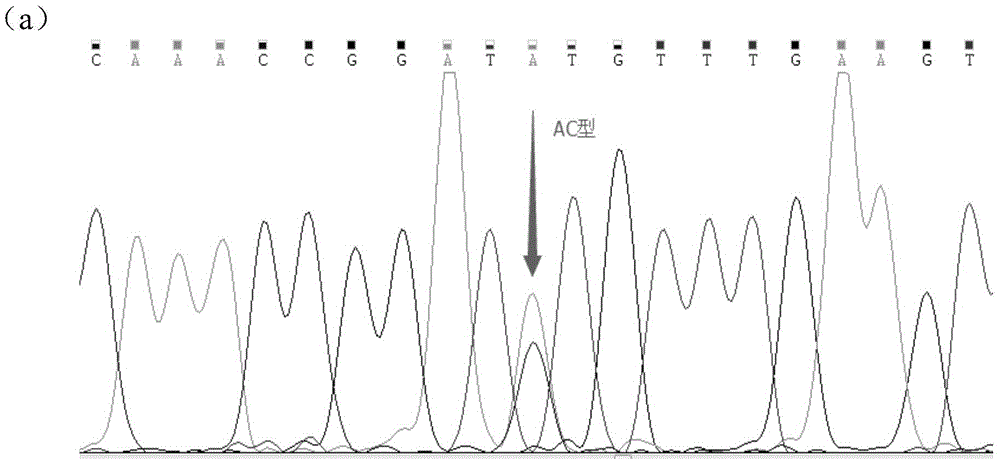
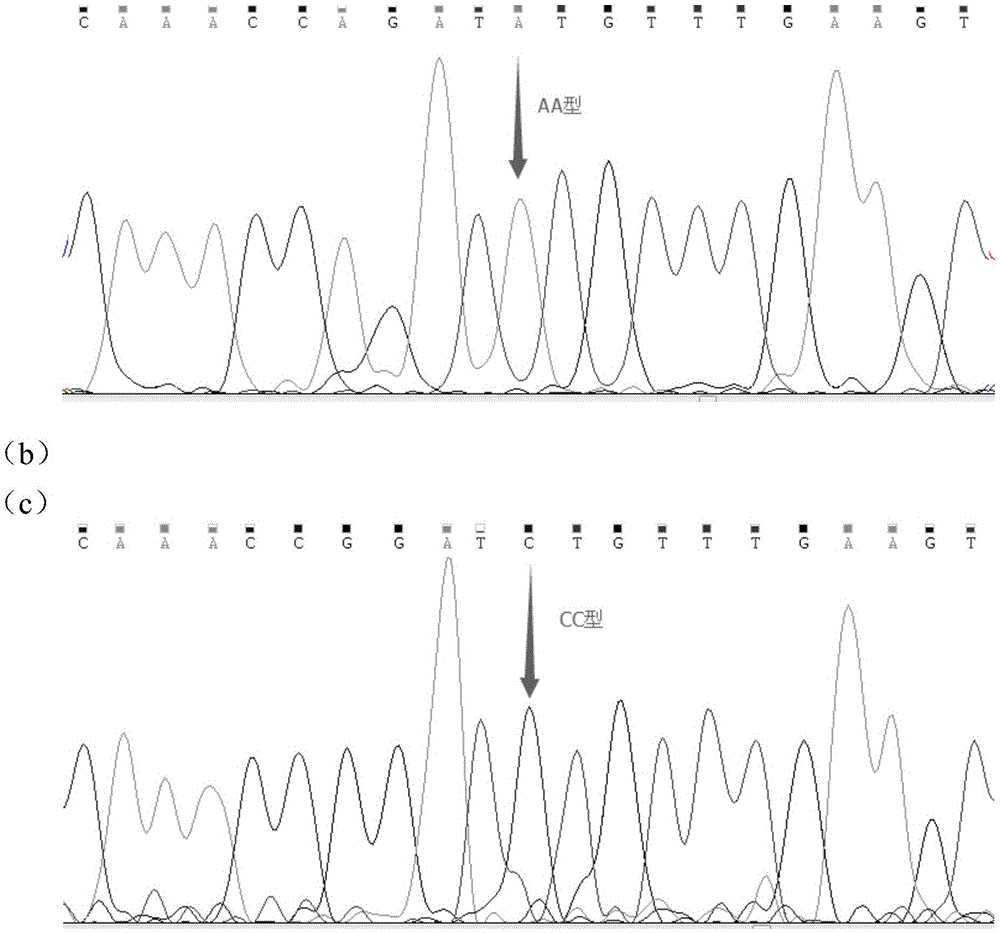
The invention discloses a molecular marker influencing the feed conversion ratio character of a pig and application, and belongs to the technical field of marker-assisted selection technologies and genetic breeding. The molecular marker is obtained through the genome-wide association study (GWAS), the nucleotide sequence of the molecular marker is shown in SEQ ID NO:1, the C>A nucleotide single-base mutation (named as g.690C>A) is located at the 690bpth position of the genomic sequence of the pig, and the feed conversion ratio character of the pig is remarkably influenced through mutation. The invention discloses preparation and application of the molecular marker.
Owner:SOUTH CHINA AGRI UNIV
Method for improving aquatic animal whole-genome selective breeding efficiency
ActiveCN110867208AImprove accuracyLow parting costClimate change adaptationProteomicsAquatic animalZoology
The invention discloses a whole-genome selective breeding method suitable for aquatic animals. Specifically, high-density SNP typing is carried out on individuals of a breeding basic group or a core group; the phenotypic value of the target character is determined; whole-genome association analysis (GWAS) is carried out by utilizing the SNP typing data and the phenotypic data to obtain a significance P value of each SNP marker; the marks are sorted from low to high according to P values; according to different traits, different marker number combinations sorted in the top are selected according to the P values, analysis is performed by adopting methods such as GBLUP, Bayes B and the like, the prediction accuracy of the different marker numbers selected according to the P values is evaluated through cross validation, and finally, the marker combination with the highest prediction accuracy is determined. SNP typing is conducted on the candidate population or the next-generation breedingpopulation through the screened optimal marker combination, breeding value prediction is conducted on the candidate population or the next-generation breeding population through adoption of GBLUP or BayesB or ssGBLUP or other methods, and therefore, the prediction accuracy can be remarkably improved.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Method for effectively anchoring candidate gene region of peanut quantitative traits
InactiveCN109694924AAchieve positioningMicrobiological testing/measurementBiotechnologyGenetic linkage disequilibrium
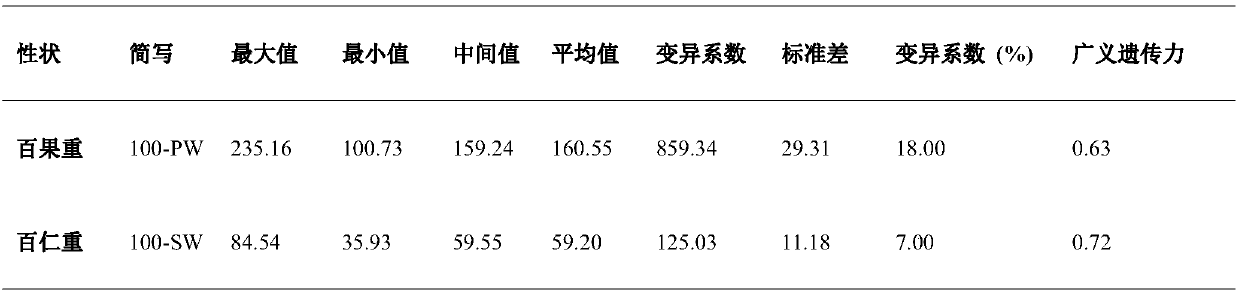
The invention relates to the field of biotechnology, and in particular to a method for effectively anchoring a candidate gene region of a quantitative trait. The method is co-localized with a genome-wide association study and a quantitative trait localization method to achieve efficient anchoring of the quantitative trait candidate gene regions. The method achieves effective anchoring of the candidate gene region of the peanut quantitative trait by co-localization with the genome-wide association study and the quantitative trait localization. The method is exemplified by quantitative granule weight, firstly, through a simplified genome sequencing technology, the genome-wide SNP marker development of 165 cultivar peanut core collections in China is carried out; based on a principle of linkage disequilibrium, combined with peanut 100-fruit weight and 100-kernel weight breeding value, the related gene regions related to peanut weight traits are associated; in the previous reports, the QTLmapping results of the grain weight traits are converted to the corresponding genomic physical locations, the co-localization region of the two methods can be obtained, and the method can effectivelyand accurately realize the localization of the genome-wide particle weight-related trait genes.
Owner:SHANDONG PEANUT RES INST
Genetic diagnosis using multiple sequence variant analysis
The present invention is in the field of nucleic acid-based genetic analysis. More particularly, it discloses novel insights into the overall structure of genetic variation in all living species. The structure can be revealed with the use of any data set of genetic variants from a particular locus. The invention is useful to define the subset of variations that are most suited as genetic markers to search for correlations with certain phenotypic traits. Additionally, the insights are useful for the development of algorithms and computer programs that convert genotype data into the constituent haplotypes that are laborious and costly to derive in an experimental way. The invention is useful in areas such as (i) genome-wide association studies, (ii) clinical in vitro diagnosis, (iii) plant and animal breeding, (iv) the identification of micro-organisms.
Owner:METHEXIS GENOMICS
Method for exploring disease-related SNP combination based on evolutionary algorithm in genome-wide association analysis data
ActiveCN109390032AHigh precisionImprove interpretabilityData visualisationProteomicsGenomeGenome-Wide Association Analysis
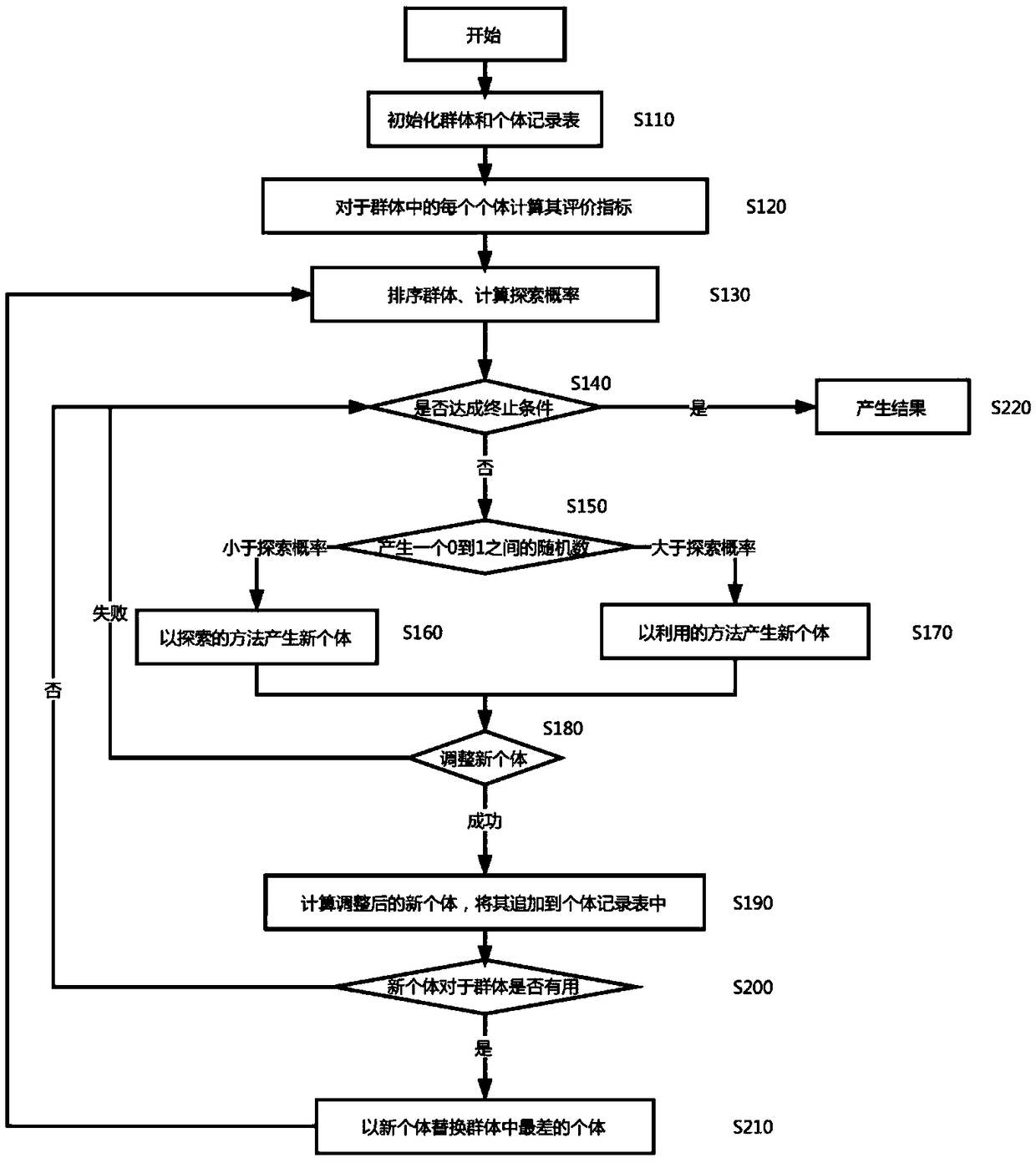
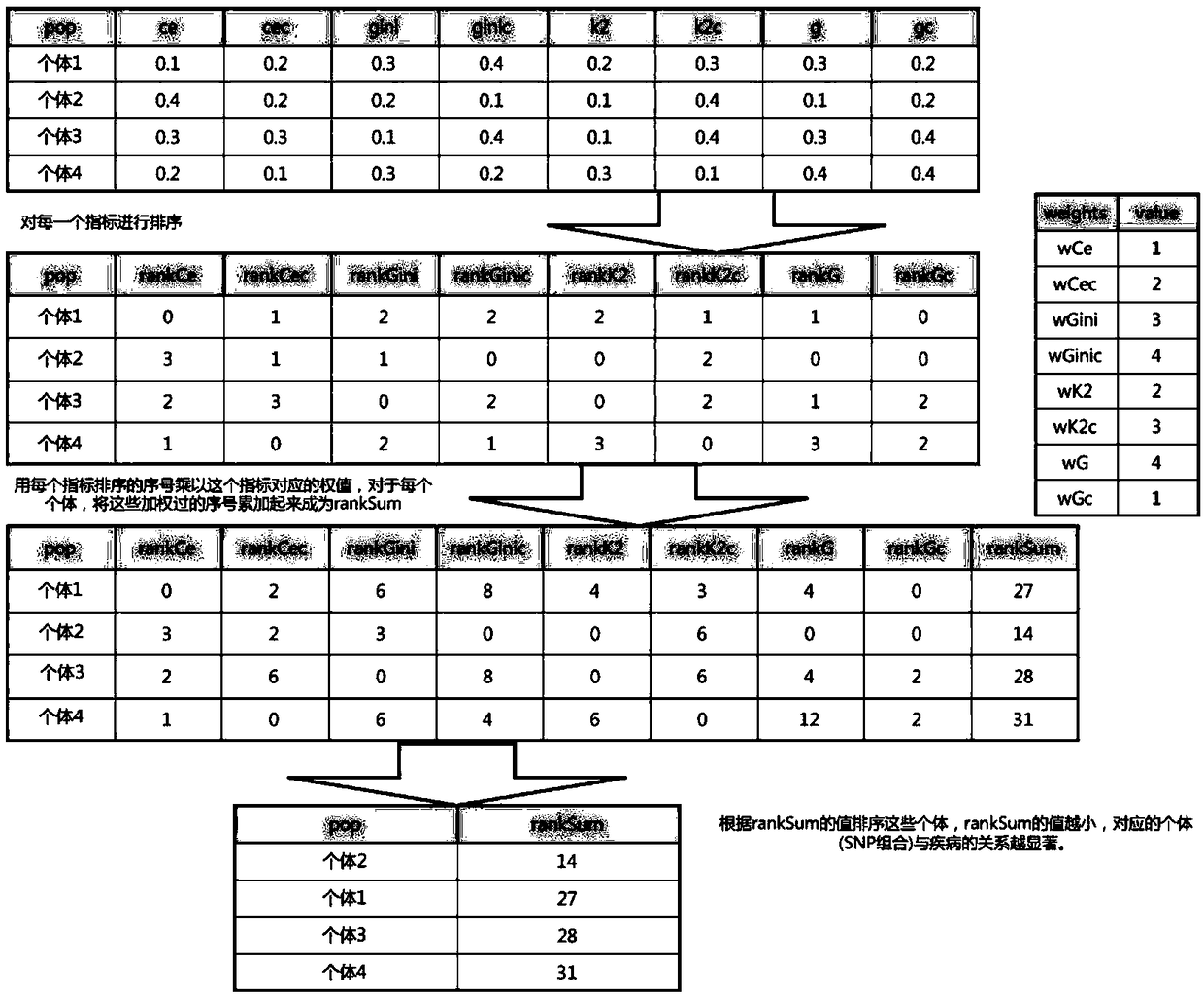
The invention discloses a method for exploring a disease-related SNP combination based on evolutionary algorithm in genome-wide association analysis data. The method comprises the following steps: step 1, initializing group and individual recording tables, calculating an evaluation index of individuals in a group, wherein the evaluation index includes ce, gini, k2, g, cec, ginic, k2c, gc; step 2,sorting and merging the evaluation indexes; step 3, determining whether evolution of the group meets a termination condition, if the termination condition is met, outputting an evolution result; step4, generating a random number between 0 and 1, and determining whether the random number is greater than exploration probability, and determining to use an exploration or utilization method to generate a new individual according to a determination result; step 5, adjusting the new individual, calculating an evaluation index of the adjusted new individual, adding the evaluation index to the individual records, and determining whether eight evaluation indexes of the new individual are greater than a maximum value of a corresponding evaluation index maintained in a current group.
Owner:JILIN UNIV
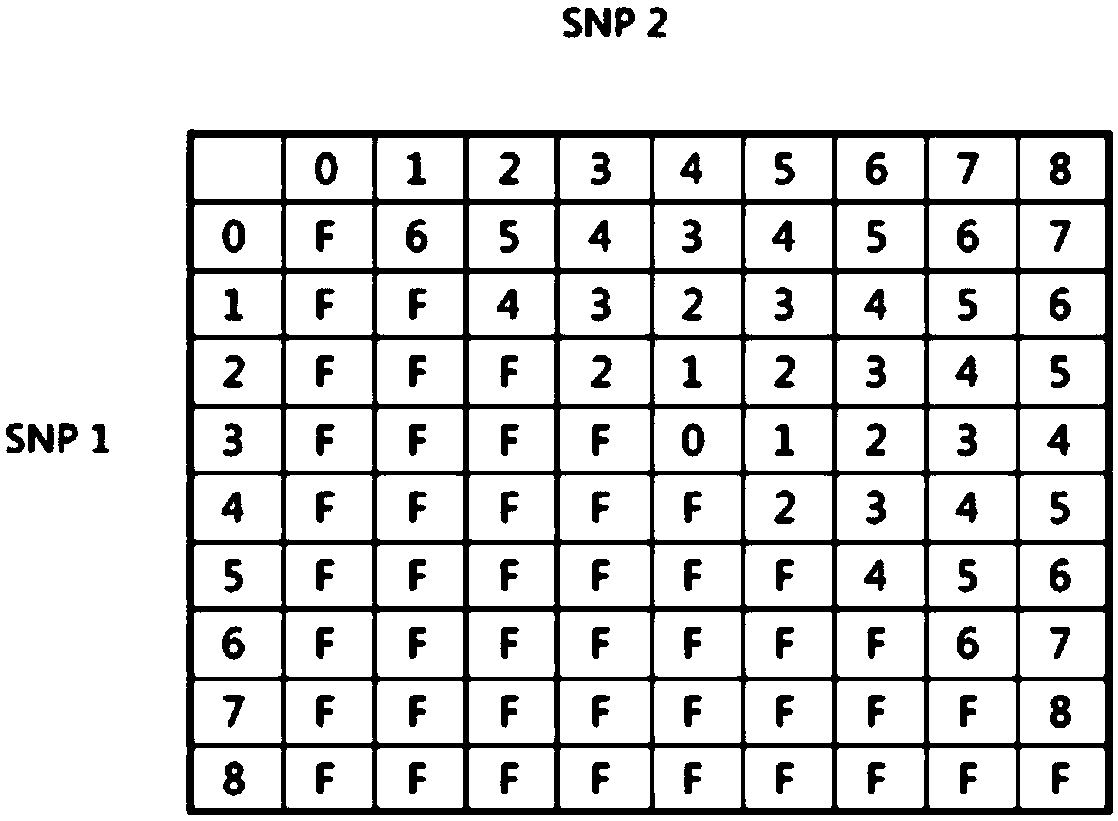
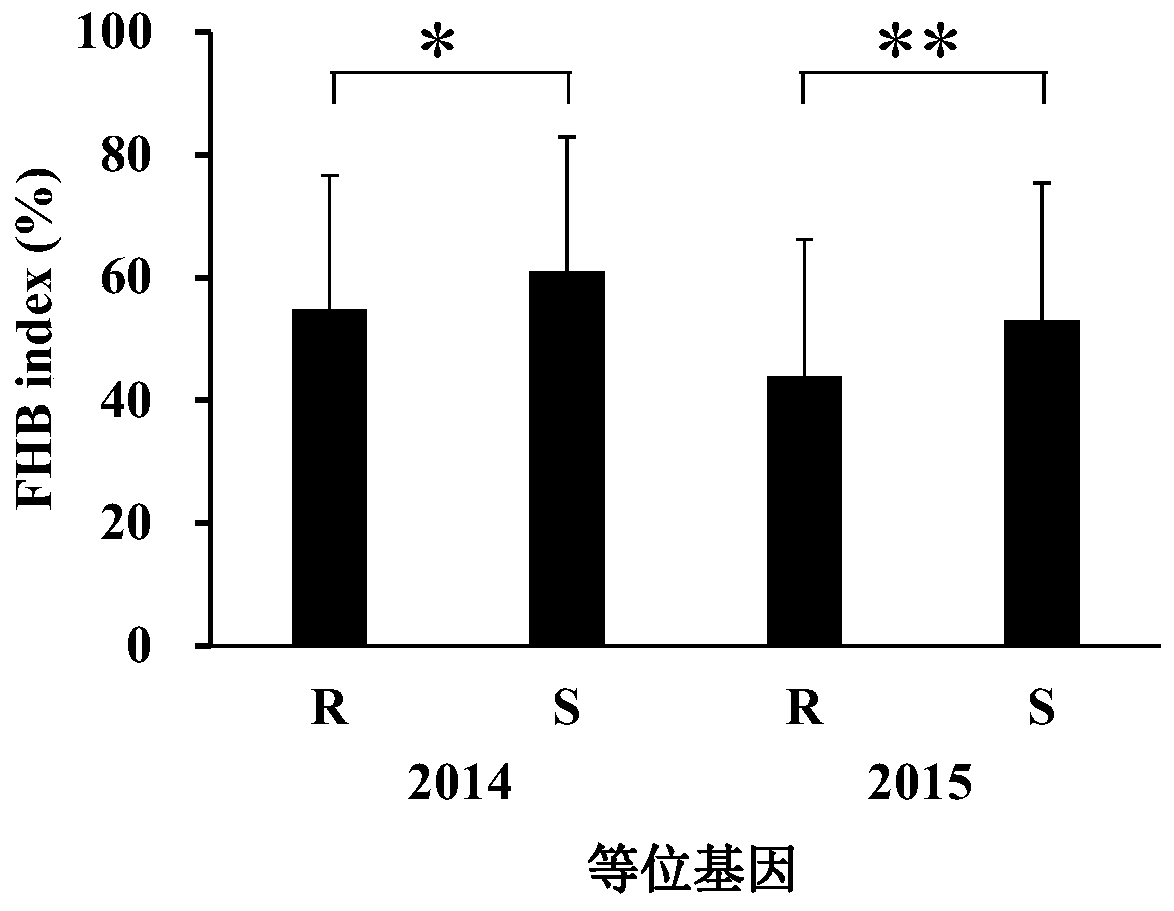
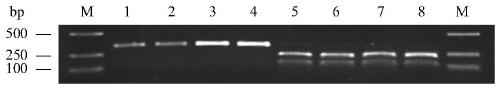
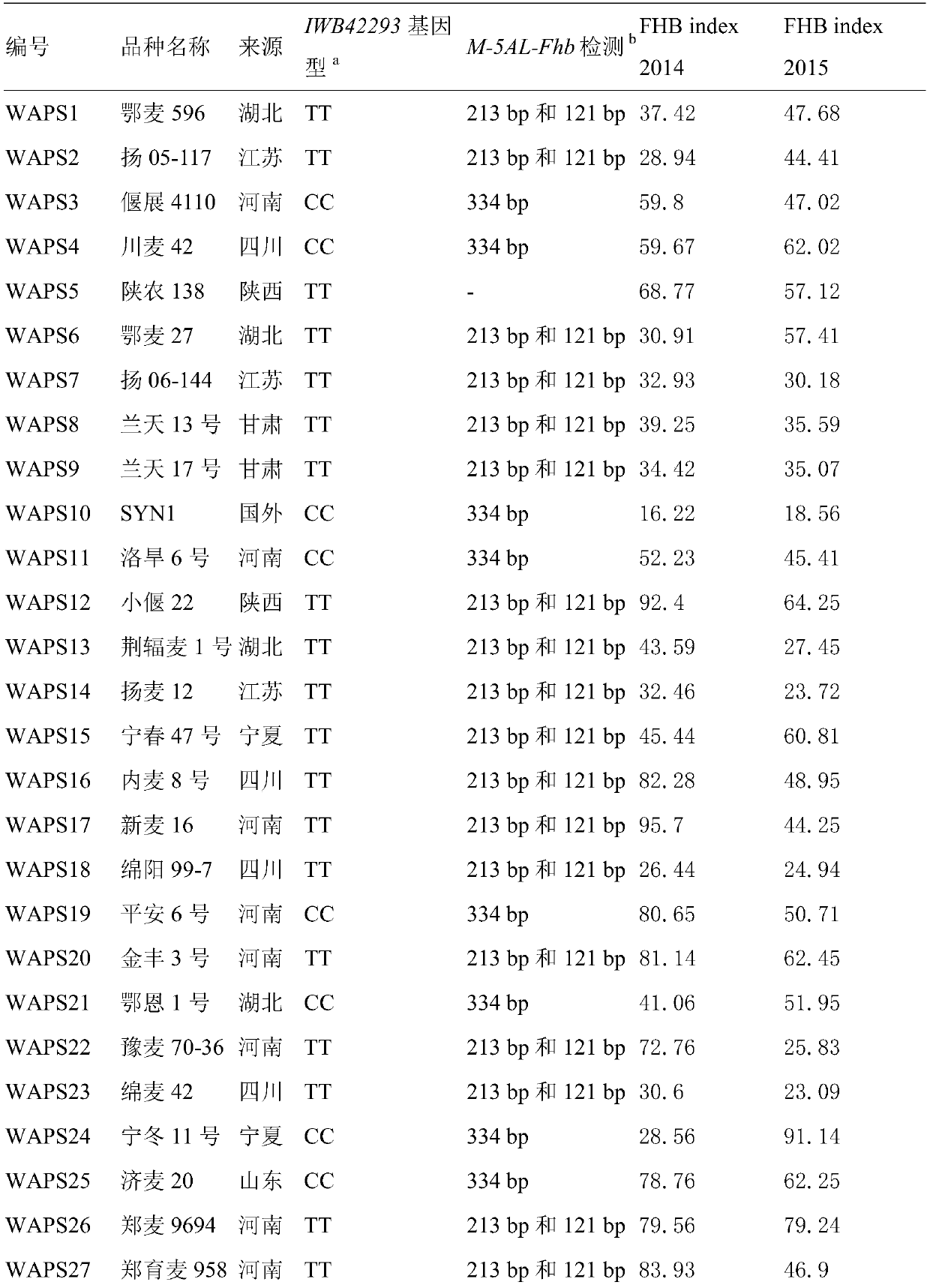
Molecular marker for detecting fusarium head blight (FHB)-resistant QTL Qfhb.hbaas-5AL and use method
ActiveCN109852719AMicrobiological testing/measurementDNA/RNA fragmentationBiotechnologyMolecular breeding
The invention discloses a molecular marker for detecting fusarium head blight (FHB)-resistant QTL Qfhb.hbaas-5AL and a use method. The invention provides an application of a substance for detecting the genotype of an SNP IWB42293 locus of wheat chromosome 5AL in identification or auxiliary identification of resistance of wheat FHB and also provides an application of the substance for detecting thegenotype of the SNP IWB42293 locus of the wheat chromosome 5AL in preparation of a product for identification or auxiliary identification of resistance of the wheat FHB. The anti-FHB locus Qfhb.hbaas-5AL located on a short arm of the wheat 5A chromosome by genome wide association study (GWAS), associated SNP IWB42293 is further converted into common PCR marked M-5AL-Fhb, and the marker can be used for detecting the genotype of the FHB-resistant QTL Qfhb.hbaas-5AL and applied to anti-FHB molecular breeding.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI +1
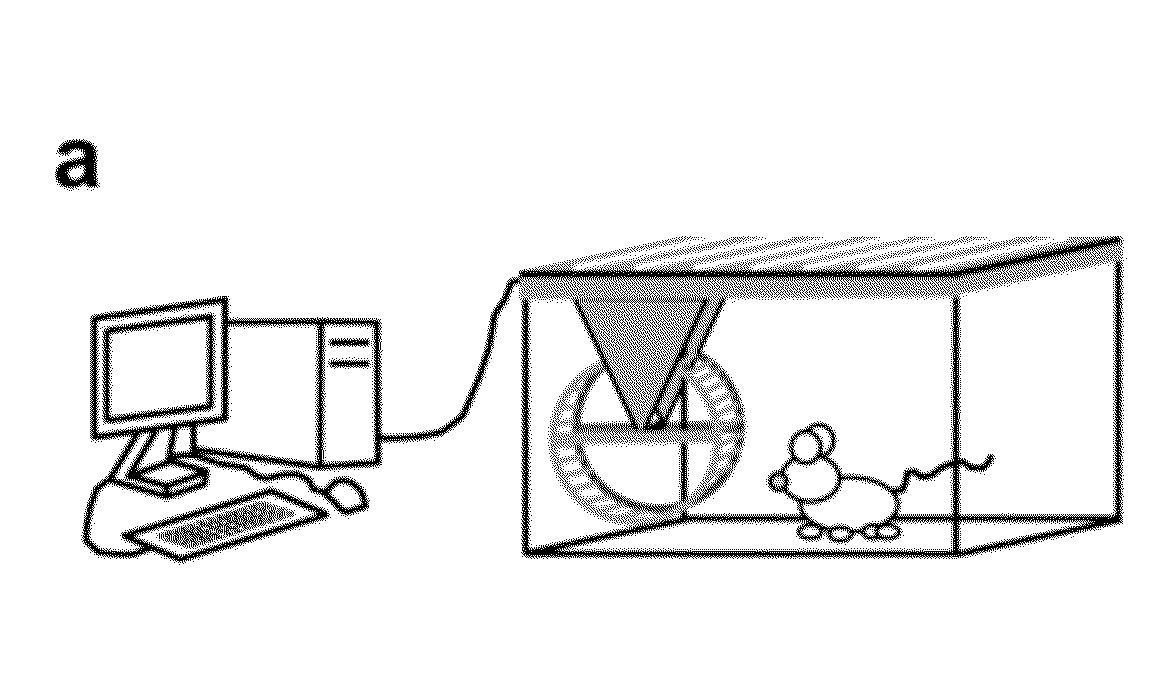
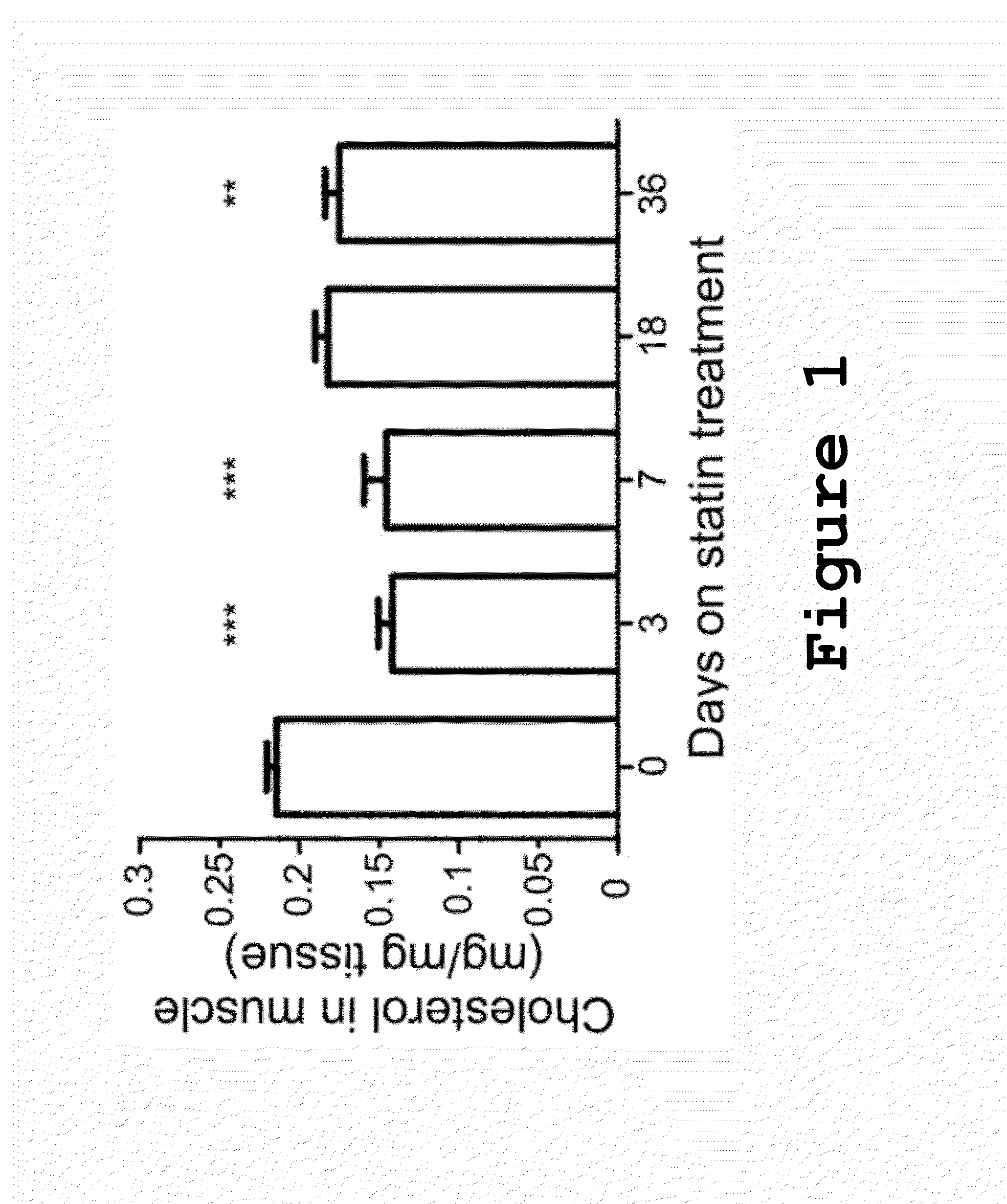
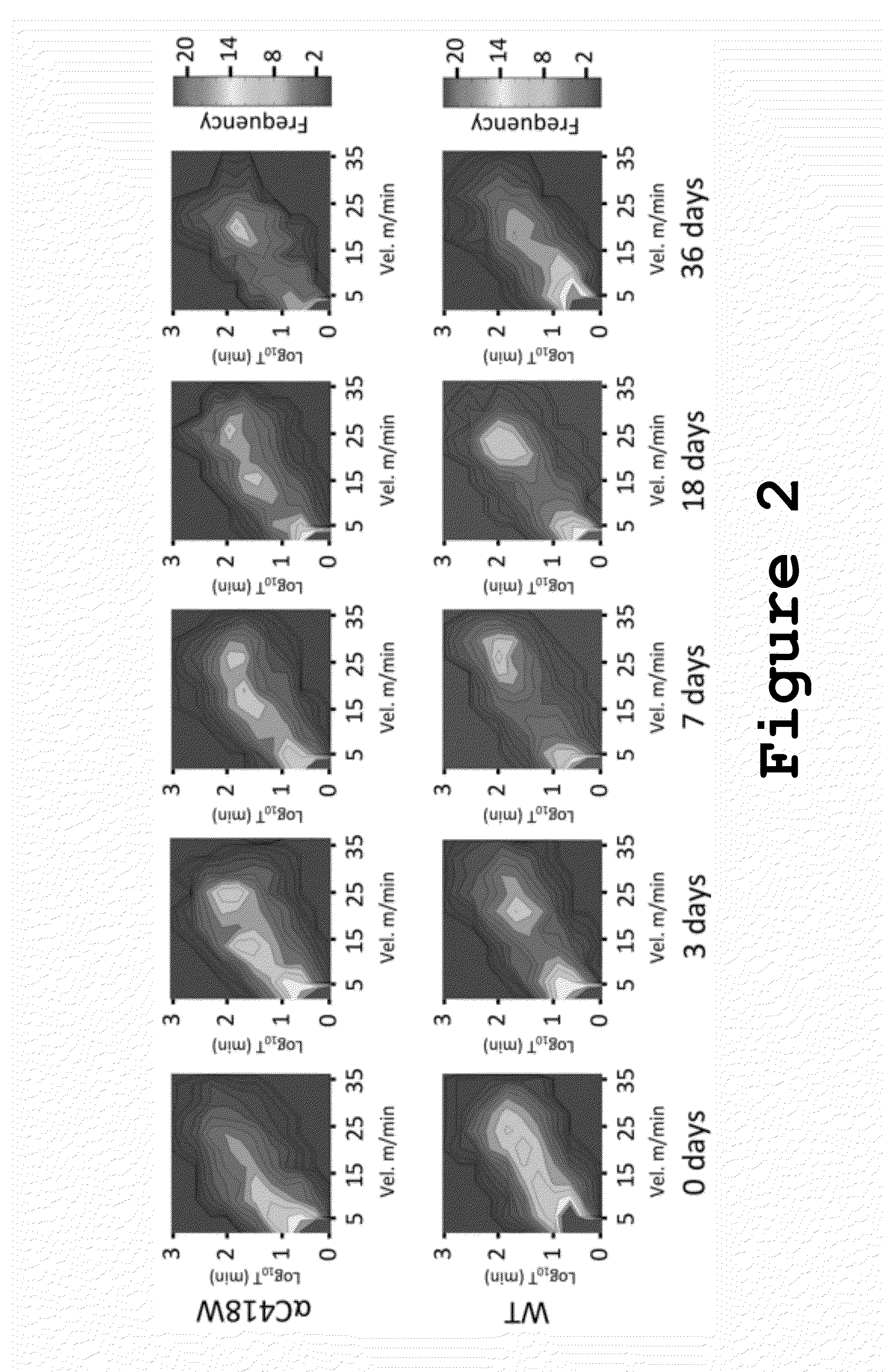
Method of Predicting Increased Risk of Suffering Statin-induced Adverse Drug Reactions
InactiveUS20140005281A1Increased riskBiocideMicrobiological testing/measurementNachr subunitSlow channel syndrome
Inhibitors of 3-hydroxy-3-methylglutaryl-coenzyme A reductase (statins) are prescribed to lower serum cholesterol levels and reduce the risk of CVD. Despite the success of statins, many patients abandon treatment owing to neuromuscular adverse drug reactions (ADRs). Genome-wide association studies have identified the single-nucleotide polymorphism (SNP) rs4149056 in the SLCO1B1 gene as being associated with an increased risk for statin-induced ADRs.By studying slow-channel syndrome transgenic mouse models, this invention determined that statins trigger ADRs in mice expressing the mutant allele of the rs137852808 SNP in the nicotinic acetylcholine receptor (nAChR) α-subunit gene CHRNA1. Mice expressing this allele show a remarkable contamination of end-plates with caveolin-1 and develop early signs of neuromuscular degeneration upon statin treatment. The invention demonstrates that genes coding for nAChR subunits may contain variants associated with statin-induced ADRs.
Owner:LASALDE JOSE A +4
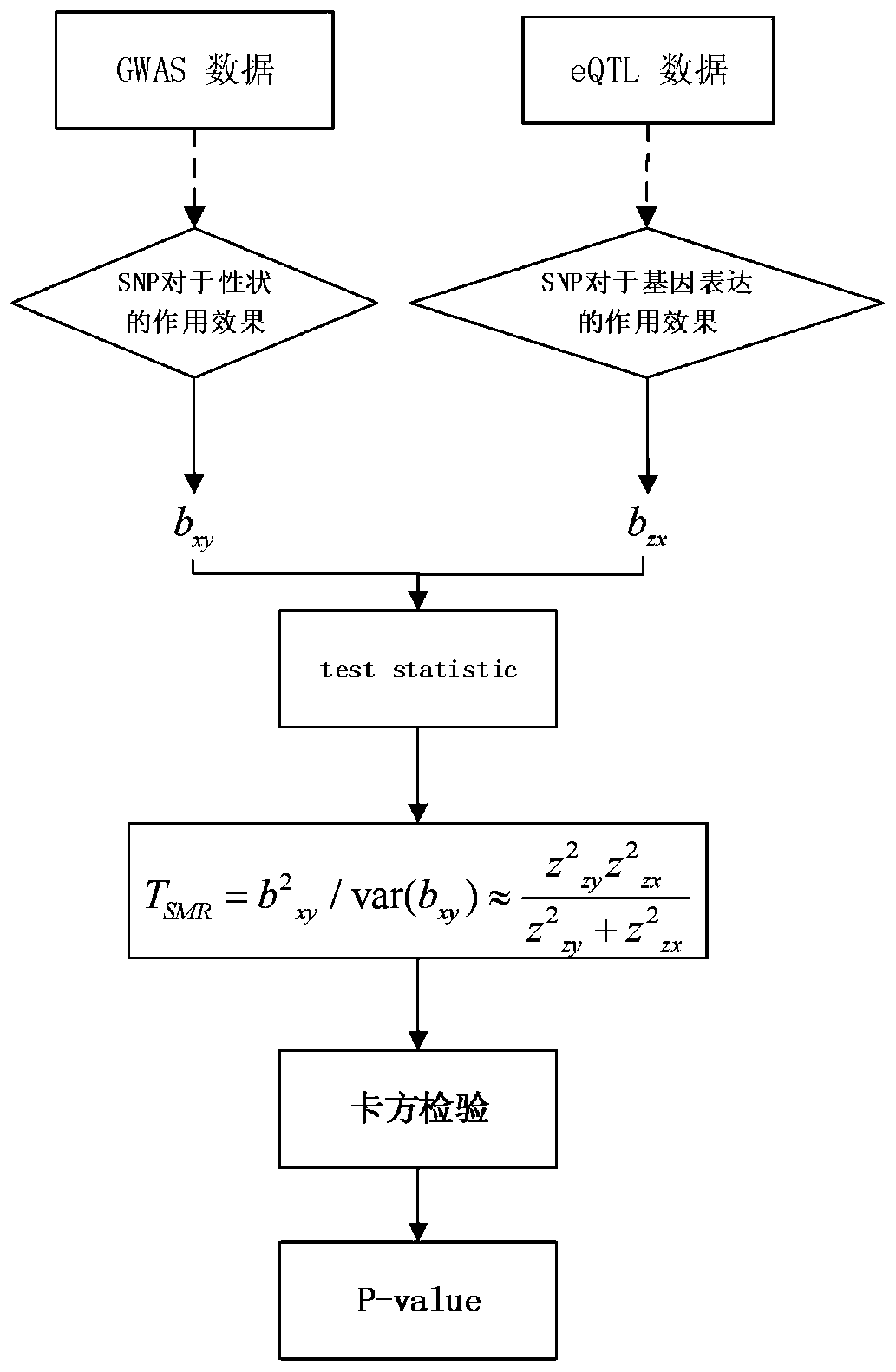
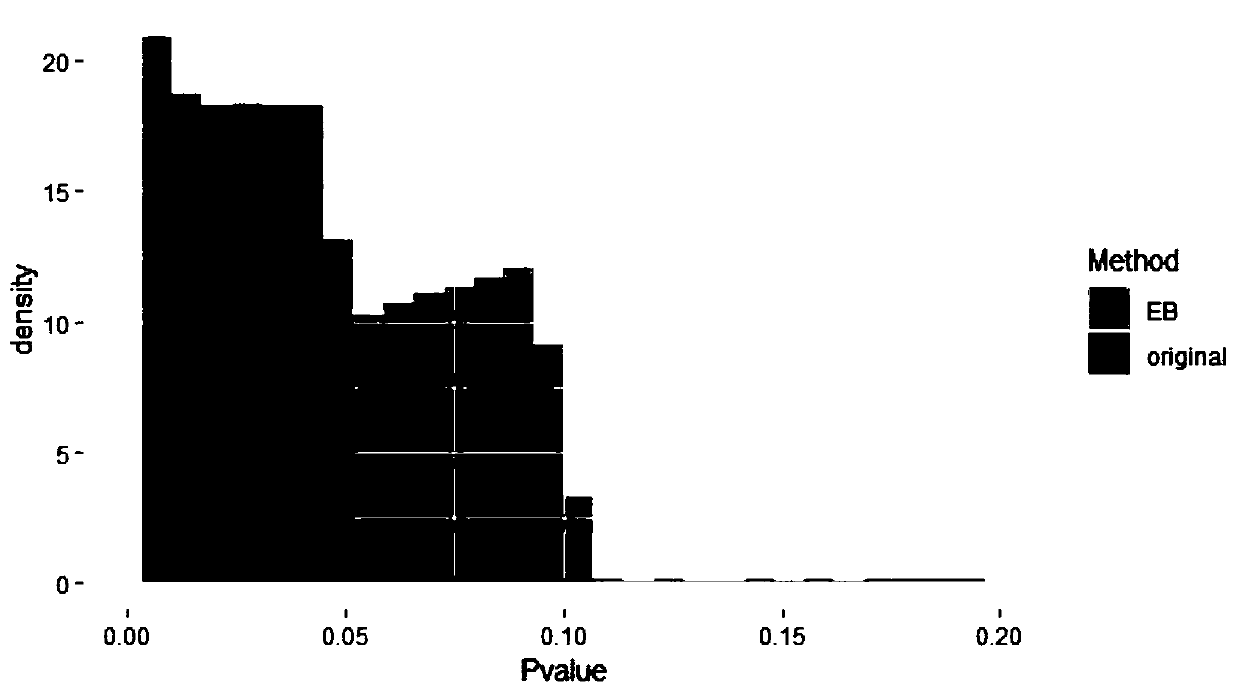
Gene recognition method based on empirical Bayesian and Mendel randomization fusion
PendingCN111180012AFast recognitionTake advantage ofBiostatisticsSequence analysisWhole Genome Association AnalysisMendelian randomization
The invention relates to a gene recognition method based on empirical Bayesian and Mendel randomization fusion. The method comprises the following steps: analyzing whole genome association analysis data by adopting empirical Bayesian meta-information to obtain an analysis result; correcting a statistical value of each SNP in the whole genome based on comprehensive hierarchical meta-information analysis of empirical Bayesian; integrating the whole genome association analysis data based on the Mendel randomization with the eQTL data and the mQTL data respectively, and obtaining a gene recognition result according to the overlapping part of the integration results of the whole genome association analysis data based on the Mendel randomization integrating with the eQTL data and the mQTL data.According to the method, the recognition speed of AD related genes can be greatly increased, existing data are fully utilized and the recognition speed of disease related genes is increased, and the research and development cost is saved; and the calculation result can screen out a great part of genes, so that a valuable research range is provided for subsequent biological experiments.
Owner:HARBIN INST OF TECH
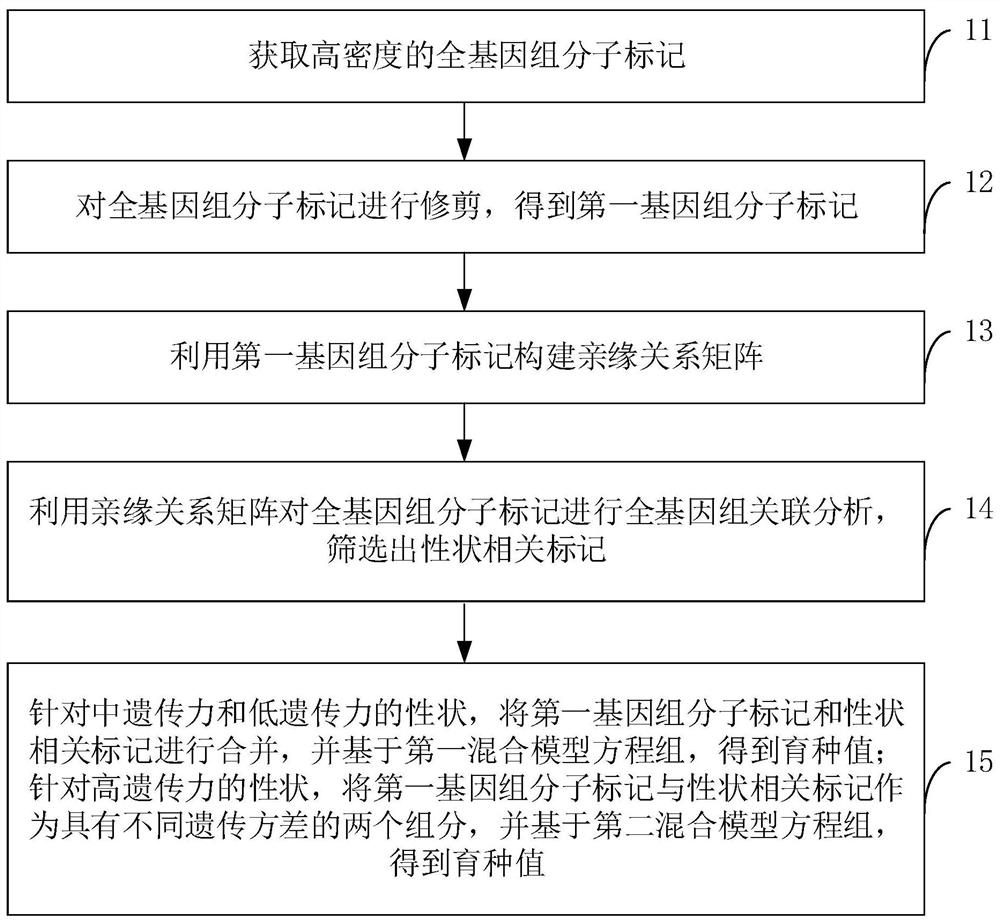
Method, device and system for estimating genome breeding value
PendingCN112712852AReduce redundancyReduce consumptionBiostatisticsHybridisationGenomeGenomewide association
The invention relates to a method, device and system for estimating a genome breeding value. The method for estimating the genome breeding value comprises the following steps: acquiring a high-density whole genome molecular marker; trimming the whole genome molecular marker to obtain a first genome molecular marker; constructing a genetic relationship matrix by utilizing the first genome molecular marker; performing whole-genome association analysis on the whole-genome molecular markers by utilizing the genetic relationship matrix, and screening out character-related markers; combining the first genome molecular marker and the trait related marker aiming at the middle genetic ability and low genetic ability traits, and obtaining a breeding value based on a first mixed model equation set; and aiming at the character with high heredity, taking the first genome molecular marker and the character related marker as two components with different heredity variances, and obtaining a breeding value based on a second mixed model equation set. Therefore, the calculation efficiency is improved, the consumption of calculation resources is reduced, the genome selection accuracy is improved, and the genetic improvement of animals and plants can be accelerated.
Owner:BGI INST OF APPLIED AGRI
Molecular marker linked with cucumis sativus peel gloss regulation gene and application
ActiveCN112592999ASolve the problem of long conventional breeding cycle and easy to be affected by the environmentReduce planting sizeFood processingMicrobiological testing/measurementBiotechnologyWhole Genome Association Analysis
The invention discloses a molecular marker linked with a cucumis sativus peel gloss regulation gene and application. The molecular marker is mutation from T to C at a 26138107 bp position on a chromosome 3 of cucumis sativus genome. 289 parts of seed resources collected by a project group are utilized, methods of whole genome association analysis (GWAS) combined with QTL positioning and the like are adopted, key variation sites influencing cucumis sativus peel gloss regulation and control are identified, and the molecular marker is developed according to the variation. One KASP molecular marker is designed, the marker is used for carrying out genotype identification on 295 single plants in a segregation population, and coincidence rate reaches 100%. Research results not only are beneficialto early identification and assisted breeding of cucumis sativus with glossy peels, but also provide a basis for map-based cloning of peel gloss regulation and control genes and analysis of molecularmechanisms of gloss regulation and control, and have a wide popularization value.
Owner:TIANJIN RES INST OF VEGETABLE
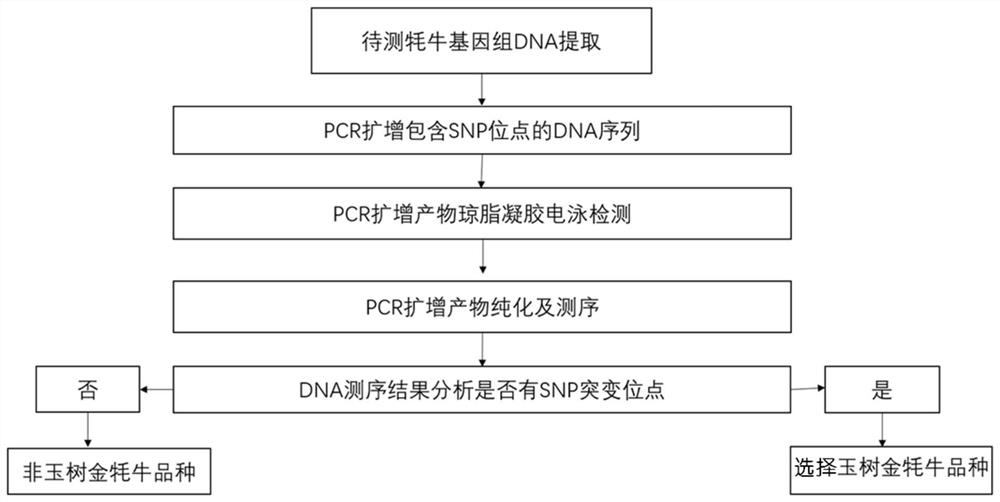
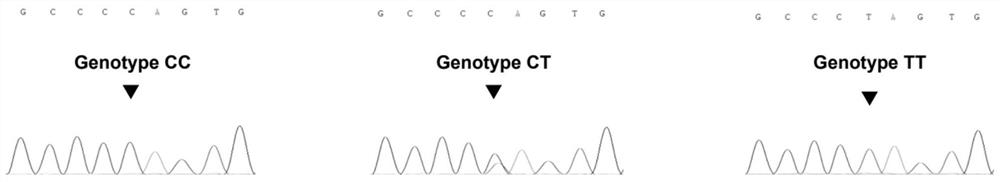
Method for identifying golden silk yak variety by using MC1R gene mononucleotide genetic marker
ActiveCN113755609AExcellent genetic resourcesQuality improvementMicrobiological testing/measurementNucleotideGenetic resources
The invention discloses a method for identifying a golden silk yak variety by using an MC1R gene mononucleotide genetic marker. According to the invention, two methods of whole-genome association analysis and whole-genome selective clearance signal calculation are utilized to analyze the whole-genome SNP of the golden-hair Yuyak, and finally a specific mutation which enables MC1R gene codons to terminate in advance on the 20# chromosome of the Yuyak is identified, so that a basis is provided for variety identification of Yuyak. Meanwhile, according to the SNP site where the mutation is located, molecular marker-assisted selective breeding can be carried out on the golden hair color character of the Yuyak on the DNA level, so that a golden yak population with excellent genetic resources is rapidly established.
Owner:NORTHWEST A & F UNIV
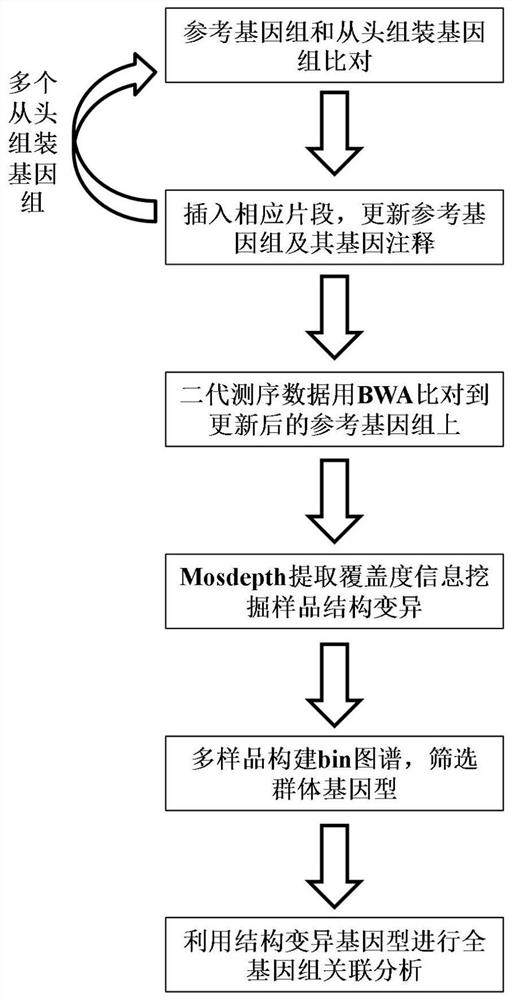
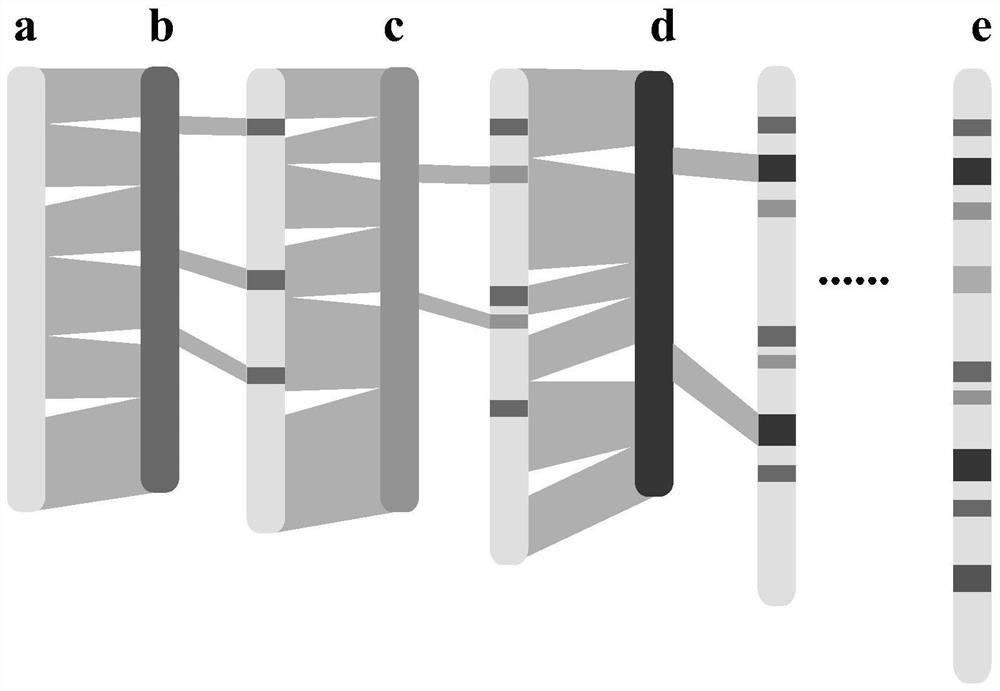
Whole genome association analysis method based on comparison of multiple genomes and next-generation sequencing data
ActiveCN113628685AImplementation of association analysisEasy to understandProteomicsGenomicsGene PositionWhole Genome Association Analysis
The invention discloses a whole genome association analysis method based on comparison of multiple genomes and next-generation sequencing data, which comprises the following steps of: 1, comparing a reference genome with a de novo assembly genome file by using comparison software; and updating the annotation gene positions or structures in the reference genomes; 3, if a plurality of de novo assembly genomes exist, sequentially iteratively updating the reference genomes; 4, comparing the next-generation sequencing data of the samples to the updated reference genome by using comparison software; 5, collecting demarcation point position information of all structural variations of all the samples into a set, constructing a population genotype; and 6, performing functional gene candidate according to the association site and the updated reference genome annotation file. According to the method, the whole genome association analysis is performed by using the structure variation genotype data.
Owner:RICE RES INST GUANGDONG ACADEMY OF AGRI SCI
Whole genome association analysis algorithm based on parent genotypes and progeny phenotypes
ActiveCN113793637ATrait high associationReduce material costsProteomicsGenomicsWhole Genome Association AnalysisGenetics
The invention discloses a whole genome association analysis algorithm based on parent genotypes and progeny phenotypes, the algorithm comprises the following steps: acquiring parent genotypes, progeny phenotypes and progeny phenotype parent information of a group to be analyzed, and establishing a combined genotype matrix of progeny according to genotypes of parent pairs, obtaining possible genotype combination information of different progenies of each SNP site and corresponding progeny group grouping phenotype data, constructing a statistical model of a progeny combination genotype and a corresponding progeny phenotype for correlation analysis, and obtaining a correlation P value of the phenotype and each SNP site; then, distinguishing correlation types of the SNP sites, calculating whether conforming to additive or complete dominant effects or not, and screening candidate sites; obtaining the average distance of the strong correlation markers according to LD attenuation of the whole genome SNP markers for screening of a final marker set.
Owner:INST OF AQUATIC LIFE ACAD SINICA
Method to predict heritable canine non-contact cruciate ligament rupture
ActiveUS20160222451A1Improve understandingHigh prevalenceMicrobiological testing/measurementAnimal husbandryCruciate ligament rupturesGenetics
Method and kits for diagnosing propensity to non-contact cranial cruciate ligament rupture (CCLR) in a dog are described. The method includes isolating genomic DNA from a dog and then analyzing the genomic DNA from step for a single nucleotide polymorphism occurring in selected loci that have been determined to be associated with the CCLR phenotype via a genome-wide association study.
Owner:WISCONSIN ALUMNI RES FOUND
Molecular marker method of clematis heat-resistant gene site qHR1
InactiveCN106434942AShorten the breeding cycleReduce breeding costsMicrobiological testing/measurementClematisHeat resistance
The invention provides a molecular marker method of a clematis heat-resistant gene site qHR1, and belongs to the field of molecular genetics. By virtue of 44 pairs of SSR molecular markers and 22 pairs of SRAP molecular markers, genotypes of 127 parts of clematis materials are determined, and in combination with heat resistance indexes, whole genome association and linkage analysis is conducted; through a GLM program and an MLM program, the clematis heat-resistant major gene site qHR1 is detected, 8.96% of phenotypic variation and 5.47% of phenotypic variation are interpreted, and an SSR molecular marker CNSSR80 is quite significantly correlated to the gene site. With the application of the molecular marker method provided by the invention, the problem of a slow progress in clematis heat-resistant breeding in China is solved, and moreover, the technical problems in an existing breeding technology which is high in breeding cost, long in duration, poor in stability and the like are solved, so that the breeding of a novel clematis heat-resistant variety in China can be accelerated.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
Schizophrenia-associated genetic loci identified in genome wide association studies and methods of use thereof
Compositions and methods for the identification of agents useful for the treatment of neurological disorders, including schizophrenia, are provided.
Owner:THE CHILDRENS HOSPITAL OF PHILADELPHIA
Molecular marking method for major QTL locus qRR5 for rooting rate in cuttage of catalpa bungei
InactiveCN105368967AShorten the breeding cycleReduce breeding costsMicrobiological testing/measurementForest industryGenotype
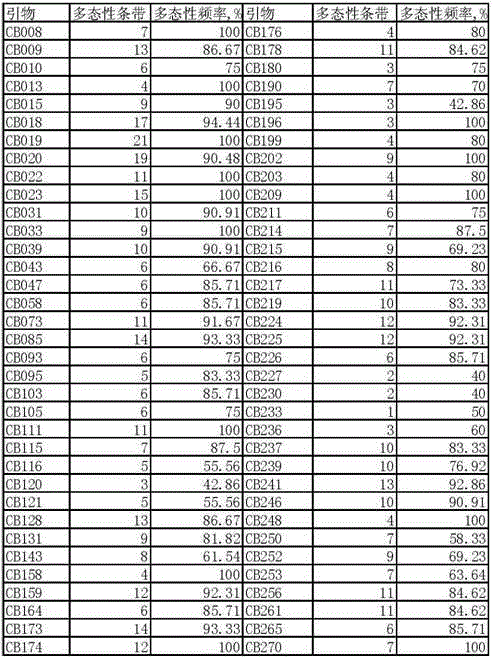
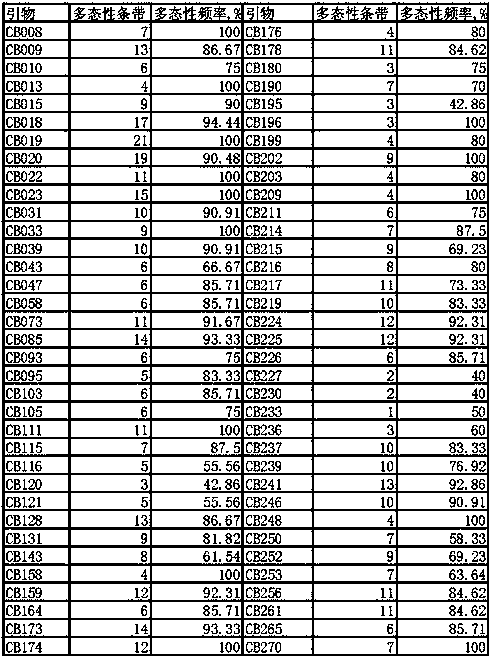
The invention provides a molecular marking method for the major QTL locus qRR5 for the rooting rate in cuttage of catalpa bungei and belongs to the field of molecular genetics. 70 pairs of SSR molecule markers are used for determining gene types of 87 parts of clonal materials of catalpa bungei, and whole genome association linkage analysis is carried out with the combination of the rooting rate in cuttage of catalpa bungei. The major QTL locus qRR5 for the rooting rate in cuttage of catalpa bungei is detected, 13.35% of phenotypic variation is explained, and the SSR molecular marker CB252 is highly remarkably associated with phenotypic variation. The molecular marking method is beneficial to solving the problem of the slow progress of catalpa bungei breeding in China and beneficial to solving the technical problem that the existing breeding technology is high in cost, long in time, poor in stability and the like when applied to increasing the rooting rate in cuttage of catalpa bungei, and breeding of new varieties of catalpa bungei in China and clone forestry development can be accelerated.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
A method for screening broodfish with high glycogen content in long oyster
ActiveCN107475414BRaw content increasedIncrease credibilityMicrobiological testing/measurementDNA/RNA fragmentationNucleotideGenetics
The invention relates to a method for screening long oyster broodstock with high glycogen content and related SNP primer pairs. Through the genome-wide association analysis of 486 individuals, 39 SNP signals significantly associated with glycogen located on chromosome 6 of Oyster chinensis were identified. A corresponding detection method was developed for a SNP site at scaffold426_295, 537 bases in this region, and the nucleotide sequence of 500 bp upstream and downstream of the site is shown in SEQ ID No.1. At the same time, compared with CC genotype individuals, the glycogen content of TT genotype individuals at this site was significantly increased by 6.2%. The TT genotype broodstock at this site was screened to guide oyster breeding. The beneficial effect of the invention is that the genotype identification of parent shellfish can be carried out before seed breeding, the TT genotype parent shellfish at this site can be screened, and the glycogen content of progeny can be increased. The results of this study are based on the SNP markers obtained from the genome-wide association analysis of the wild population, and the effect is more stable.
Owner:INST OF OCEANOLOGY - CHINESE ACAD OF SCI
Molecular marker method of main effect qtl locus qrr5 of rooting rate of catalpa cuttage
InactiveCN105368967BShorten the breeding cycleReduce breeding costsMicrobiological testing/measurementDNA/RNA fragmentationGenotypeMolecular genetics
The invention provides a molecular marking method for the major QTL locus qRR5 for the rooting rate in cuttage of catalpa bungei and belongs to the field of molecular genetics. 70 pairs of SSR molecule markers are used for determining gene types of 87 parts of clonal materials of catalpa bungei, and whole genome association linkage analysis is carried out with the combination of the rooting rate in cuttage of catalpa bungei. The major QTL locus qRR5 for the rooting rate in cuttage of catalpa bungei is detected, 13.35% of phenotypic variation is explained, and the SSR molecular marker CB252 is highly remarkably associated with phenotypic variation. The molecular marking method is beneficial to solving the problem of the slow progress of catalpa bungei breeding in China and beneficial to solving the technical problem that the existing breeding technology is high in cost, long in time, poor in stability and the like when applied to increasing the rooting rate in cuttage of catalpa bungei, and breeding of new varieties of catalpa bungei in China and clone forestry development can be accelerated.
Owner:INST OF BOTANY JIANGSU PROVINCE & CHINESE ACADEMY OF SCI
Analysis method and device for bone density character heritability
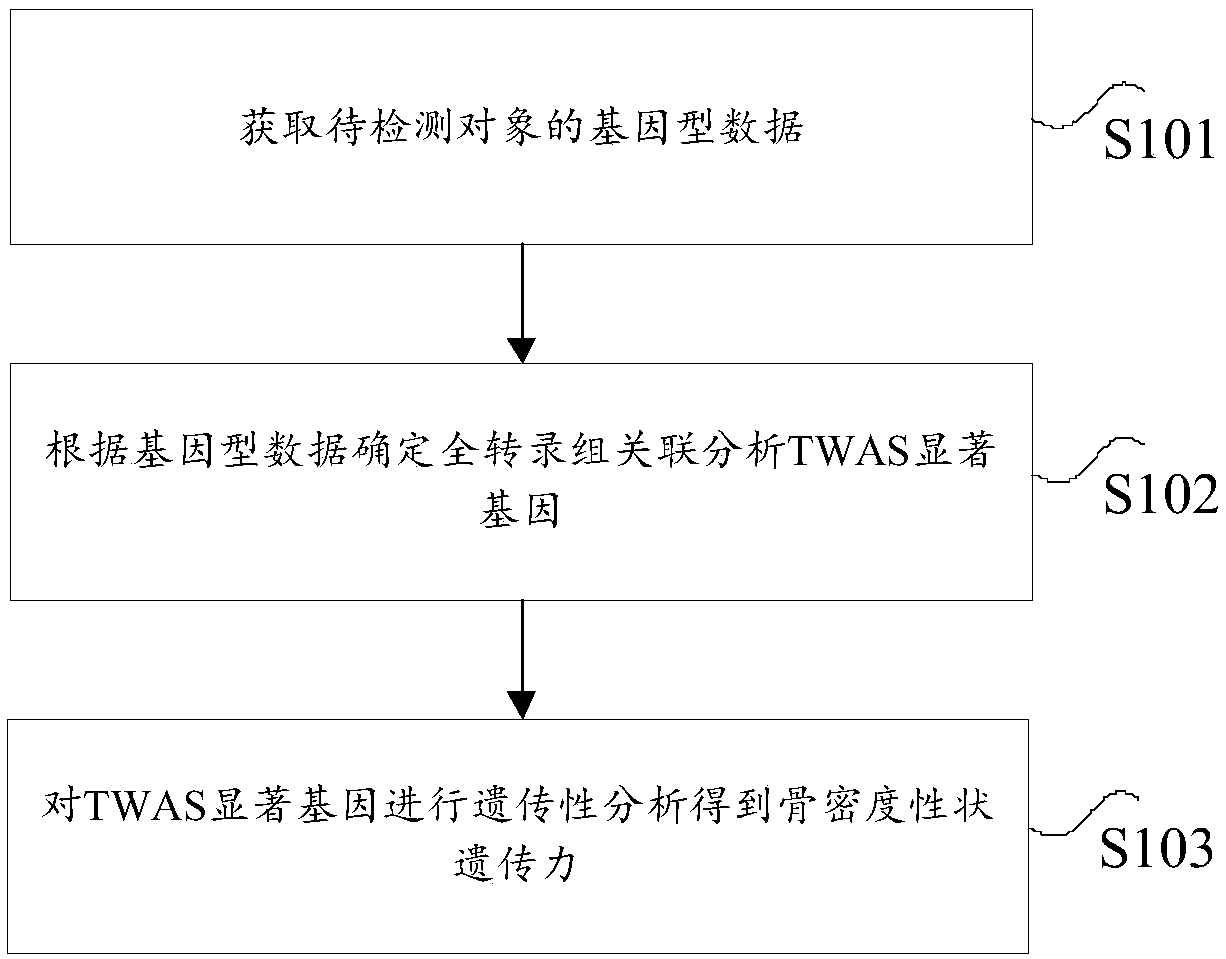
ActiveCN109637582AReduce experimental analysisHigh precisionBiostatisticsProteomicsBone densityCorrelation analysis
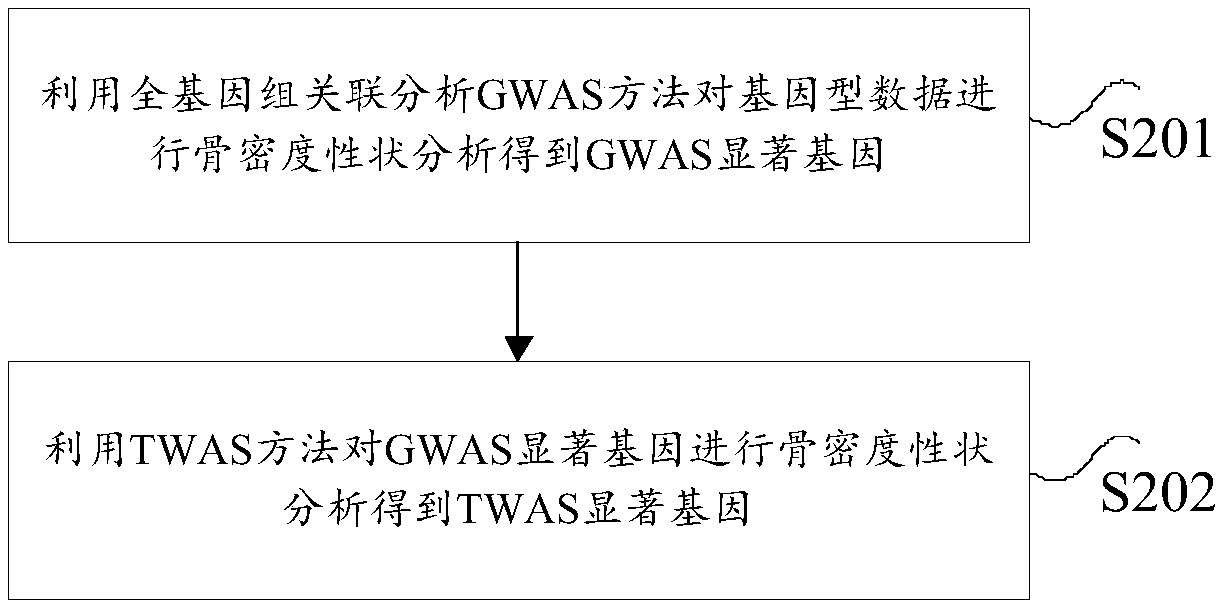
The invention provides an analysis method and device for bone density character heritability and relates to the technical field of heritability analysis. According to the method, gene-type data of a to-be-detected object is acquired; according to the gene-type data, a full-transcriptome correlation analysis TWAS obvious gene is determined; the TWAS obvious gene is subjected to hereditary analysisto obtain the bone density character heritability, a full-transcriptome management analysis method TWAS is used for screening the full-genome correlation analysis GWAS obvious gene again, the experiment analysis to the GWAS obvious gene is reduced, the accuracy of the heritability is high, and through risk grading for the heritability, the heredity interpretability is further achieved from the scientific angle.
Owner:SHENZHEN INST OF ADVANCED TECH
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com