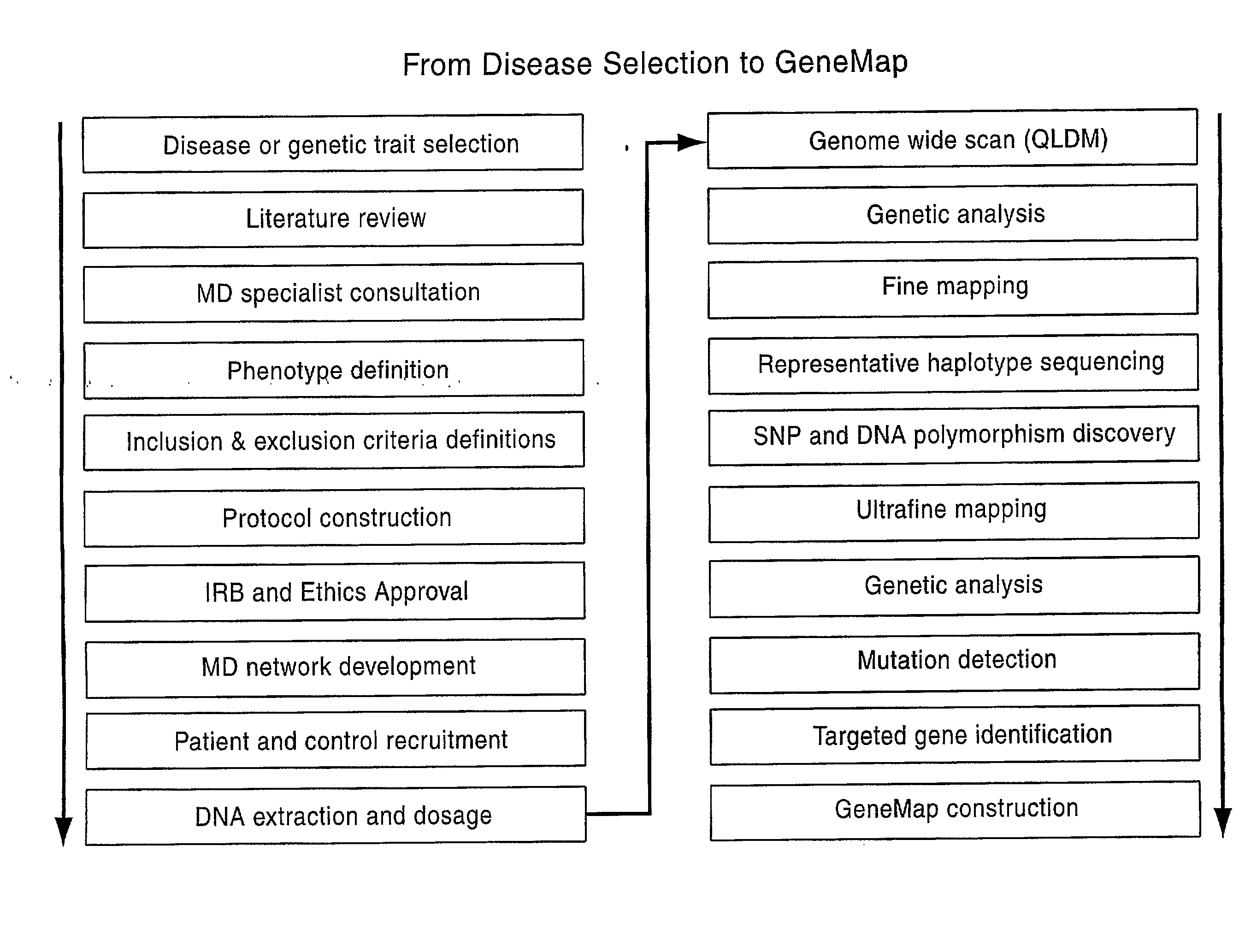
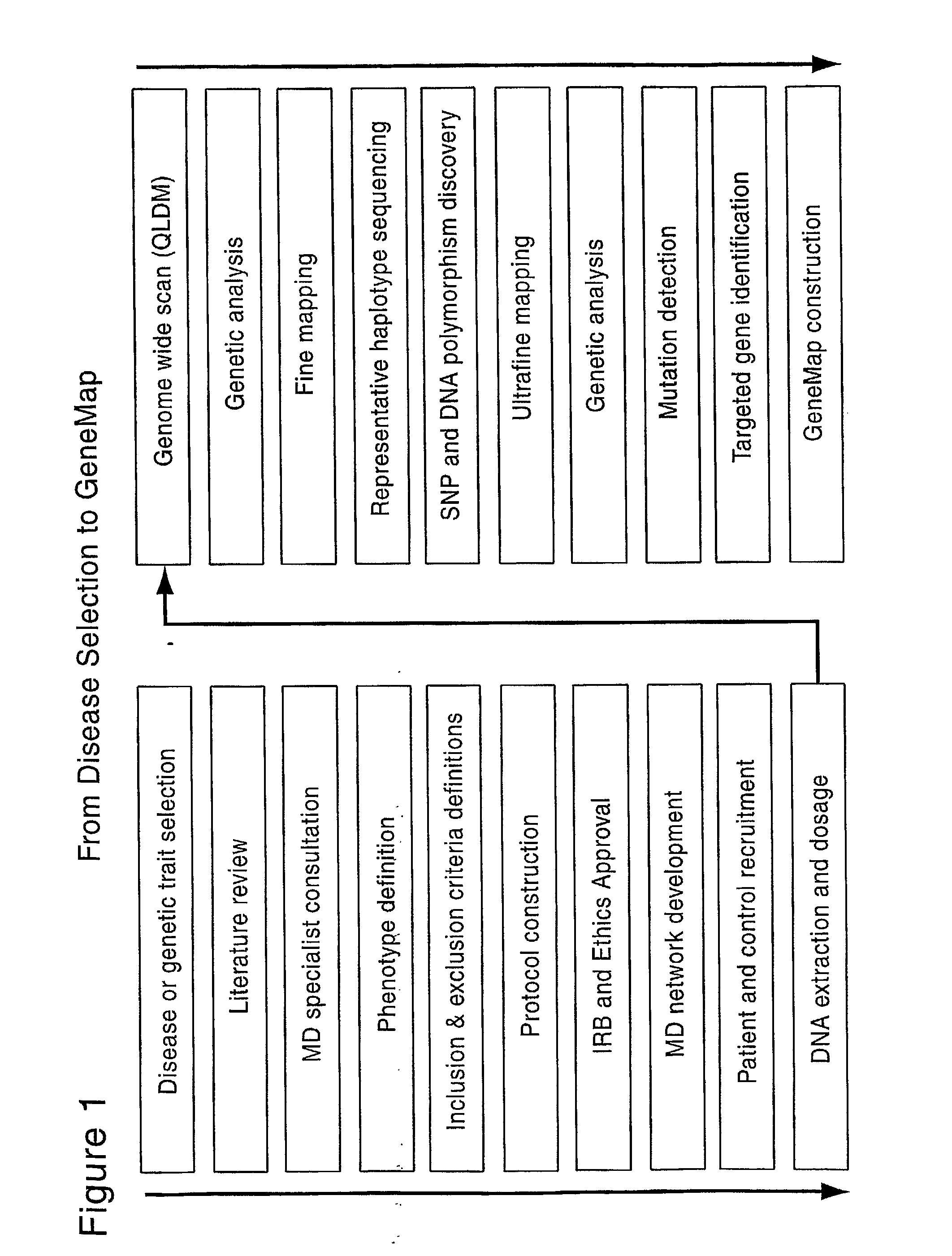
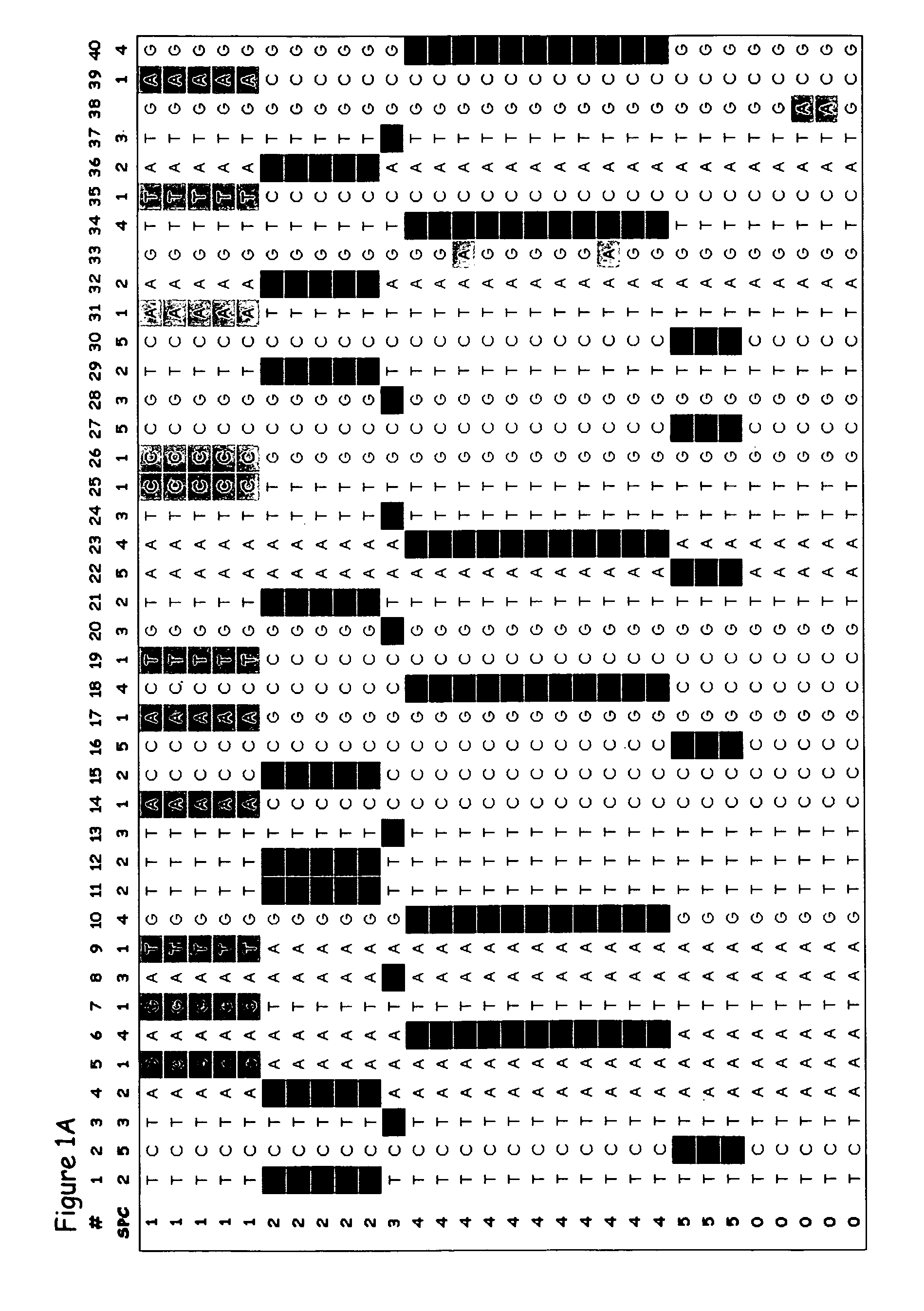
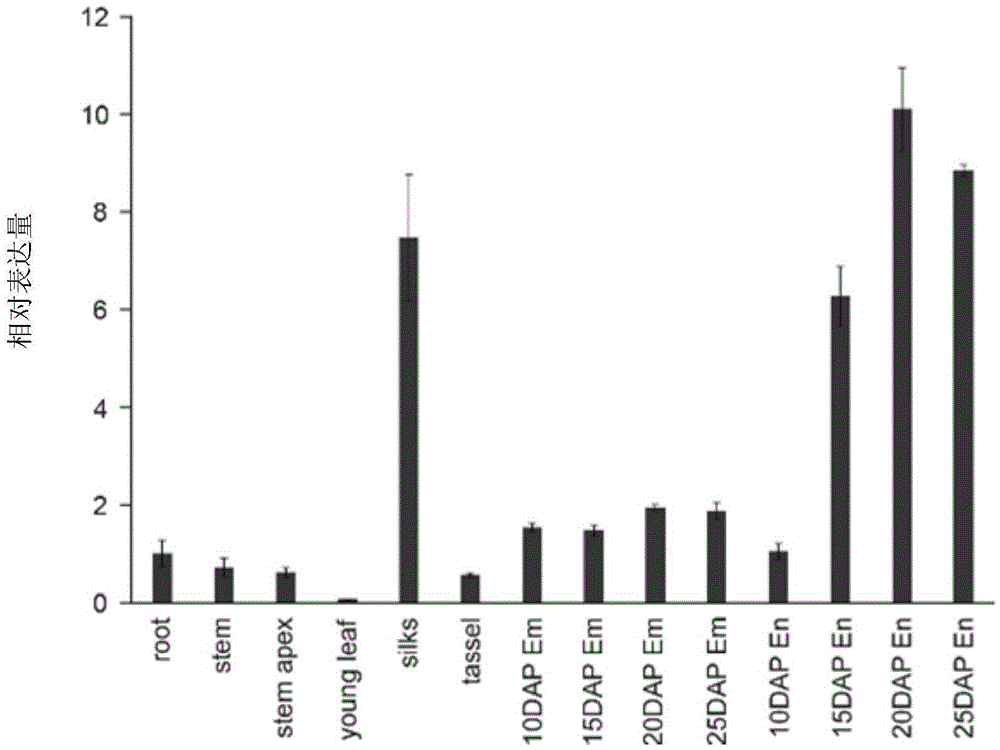
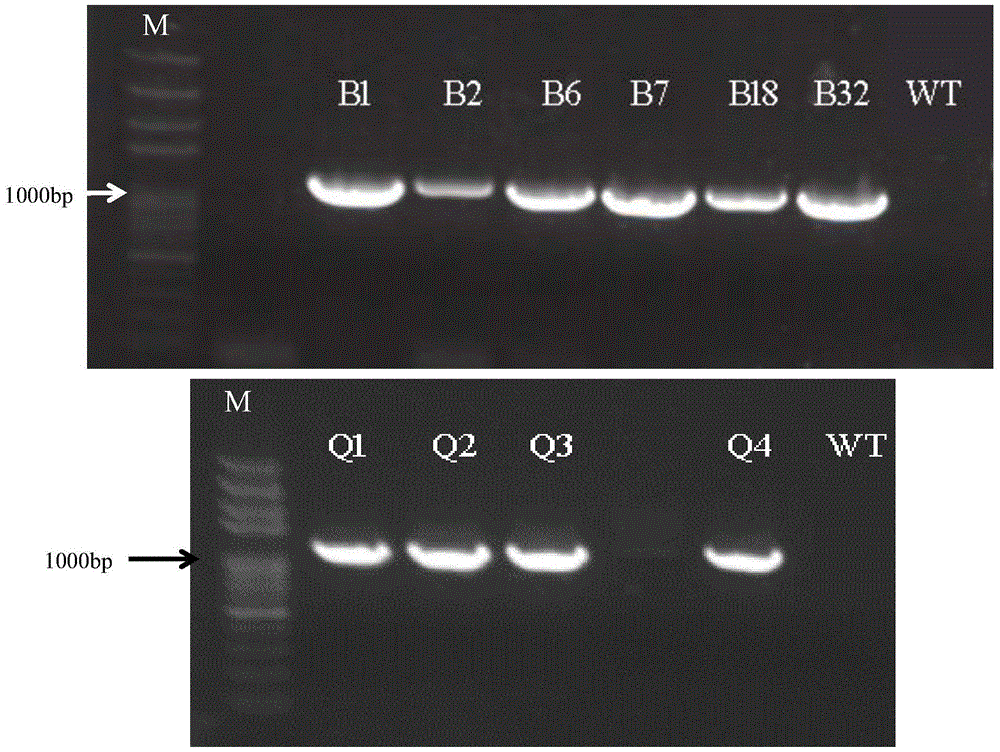
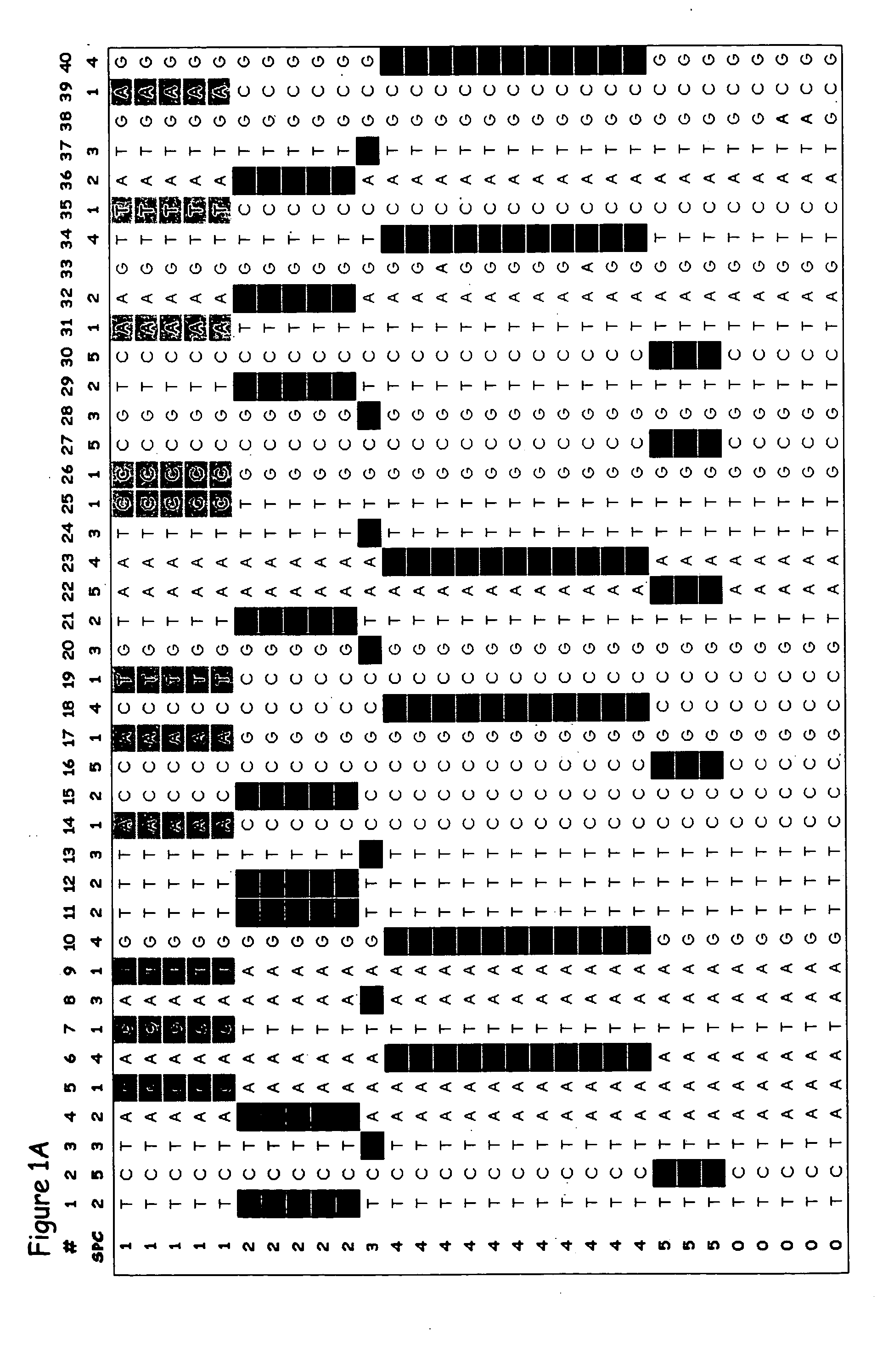
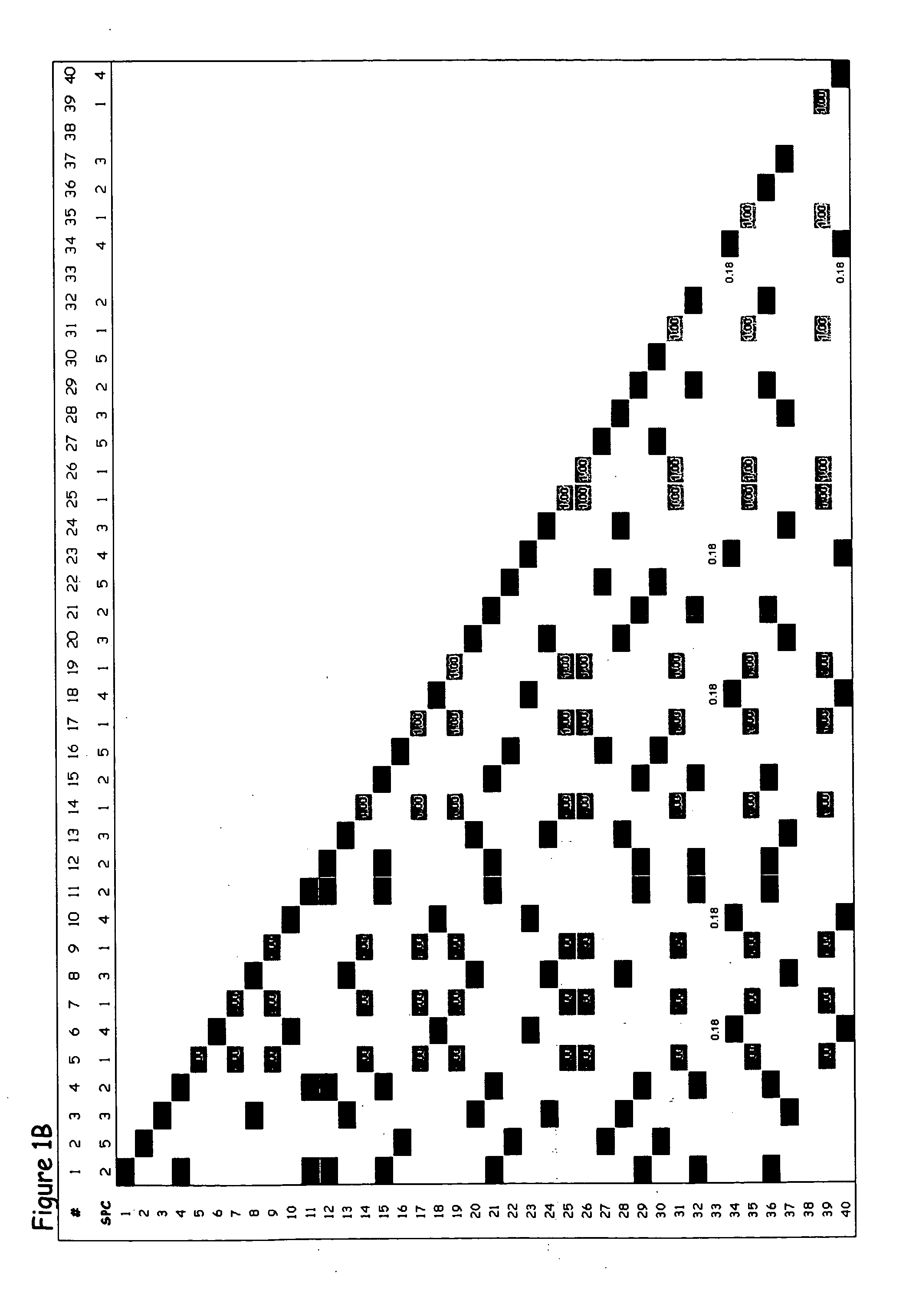
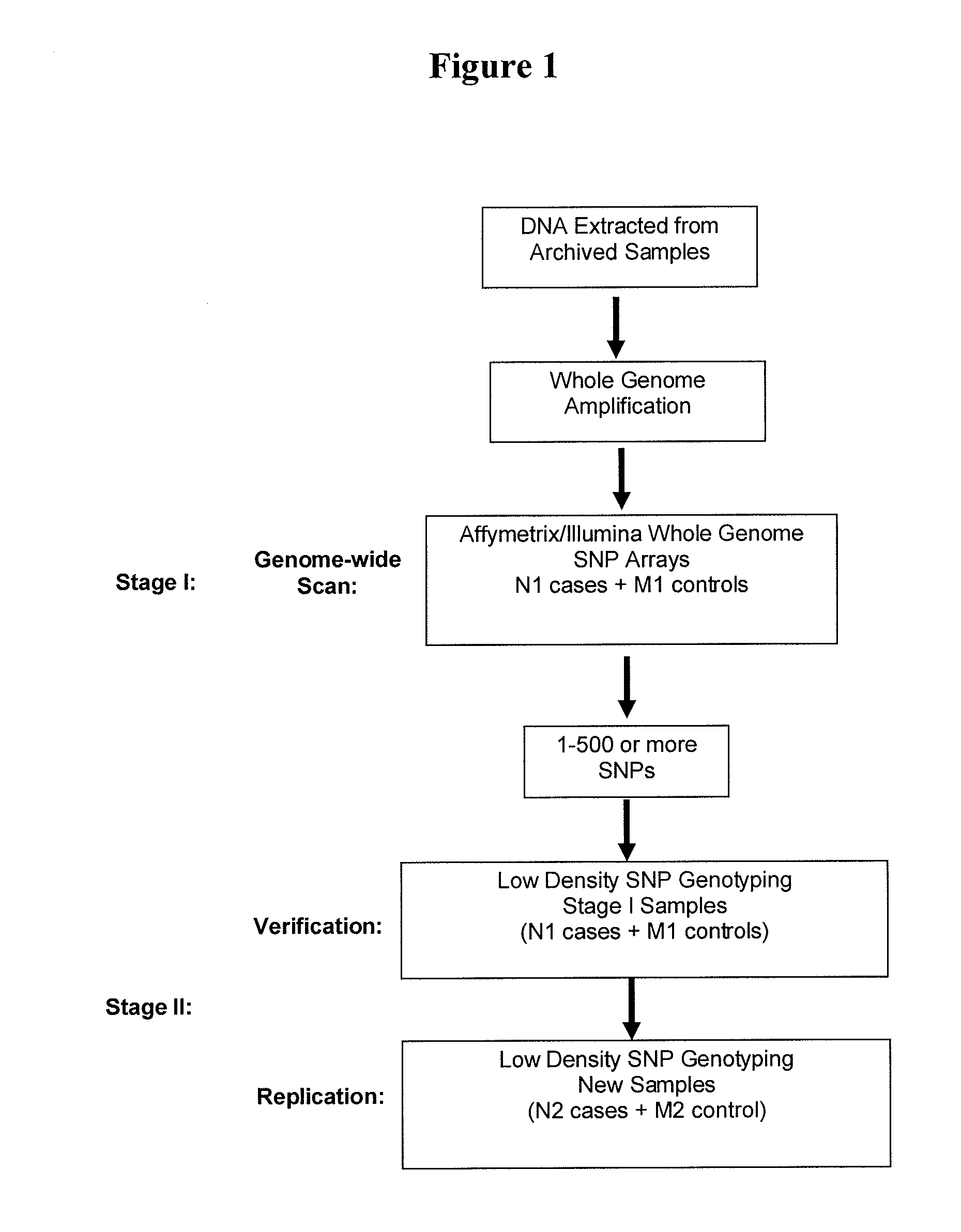
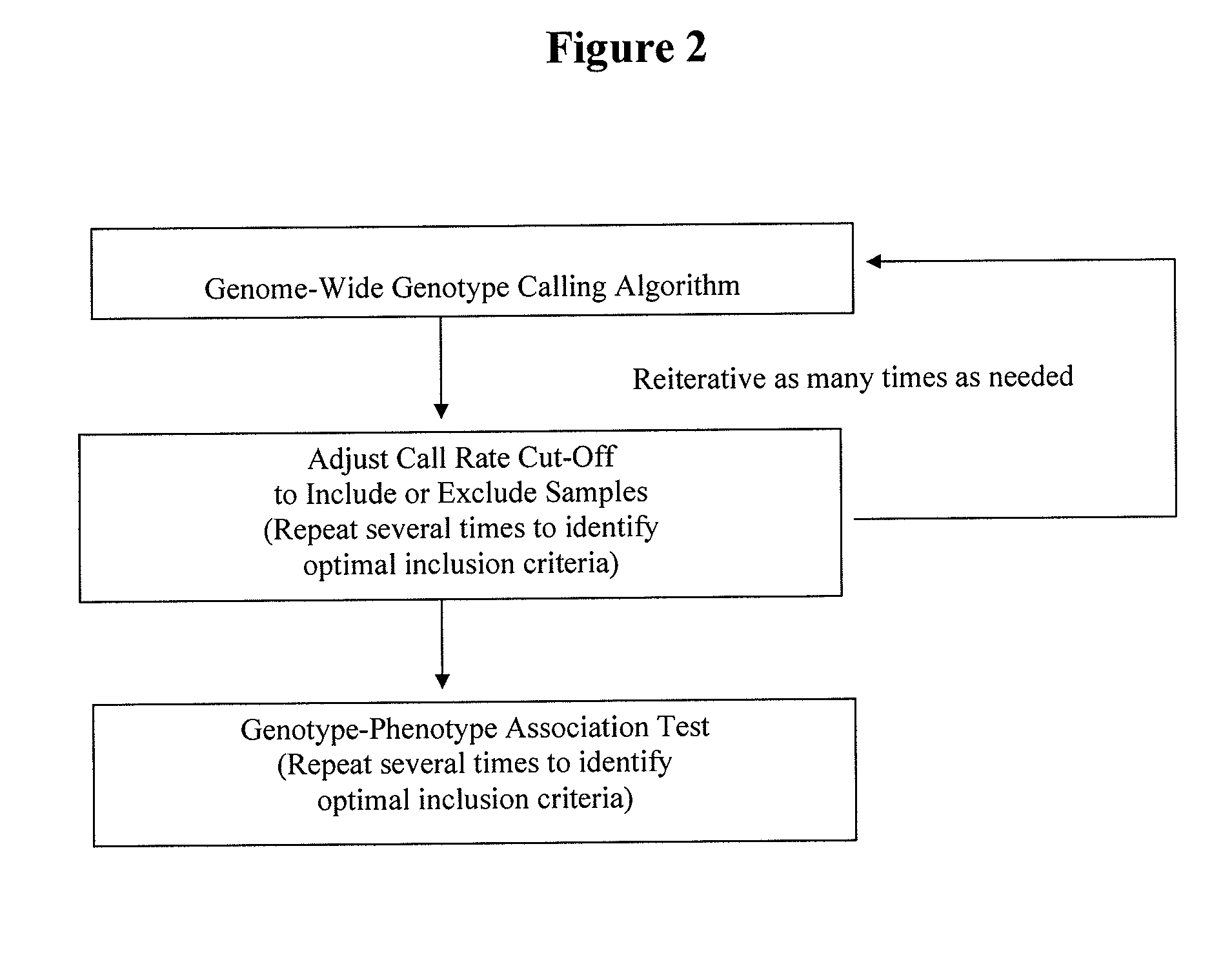
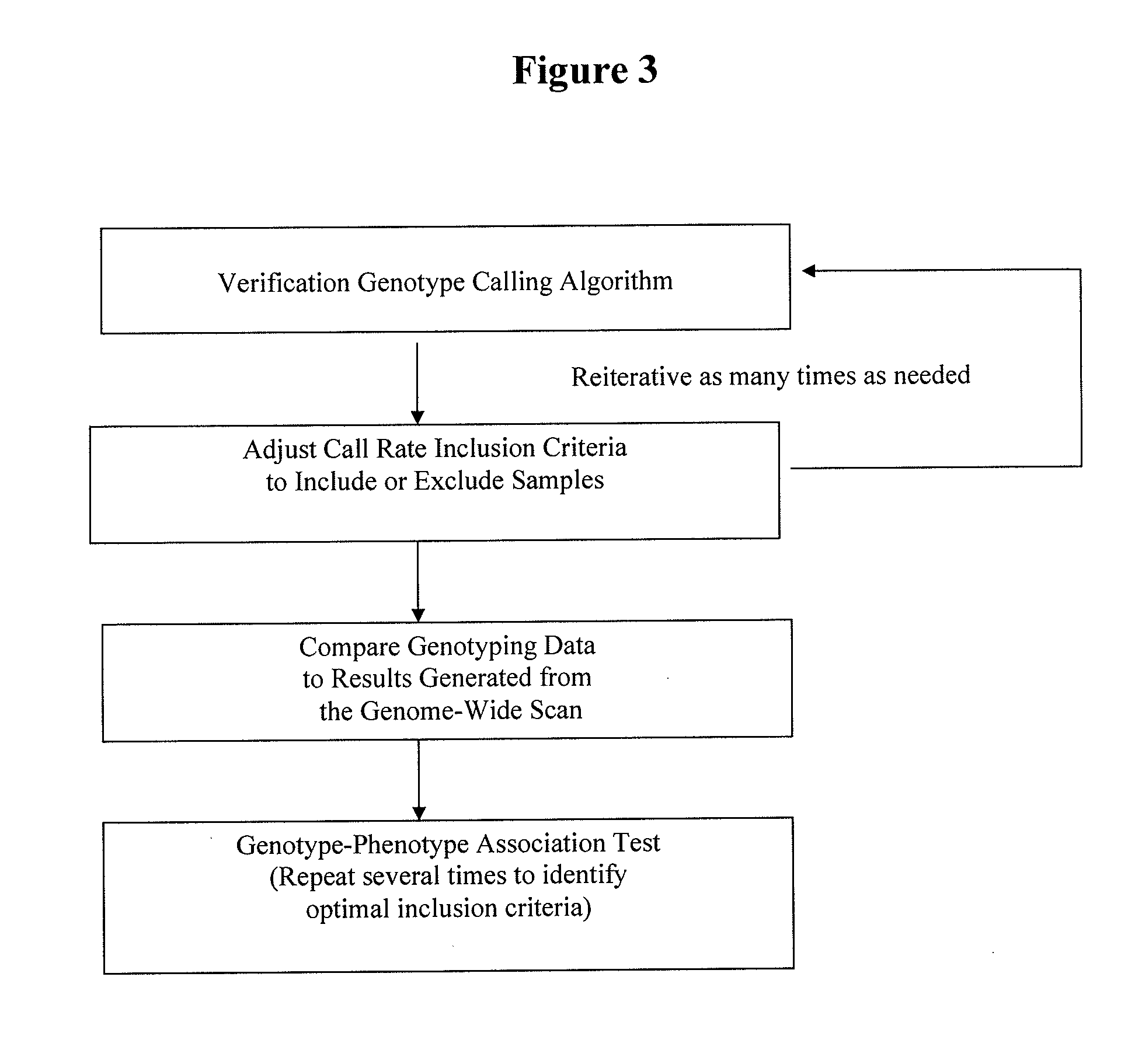
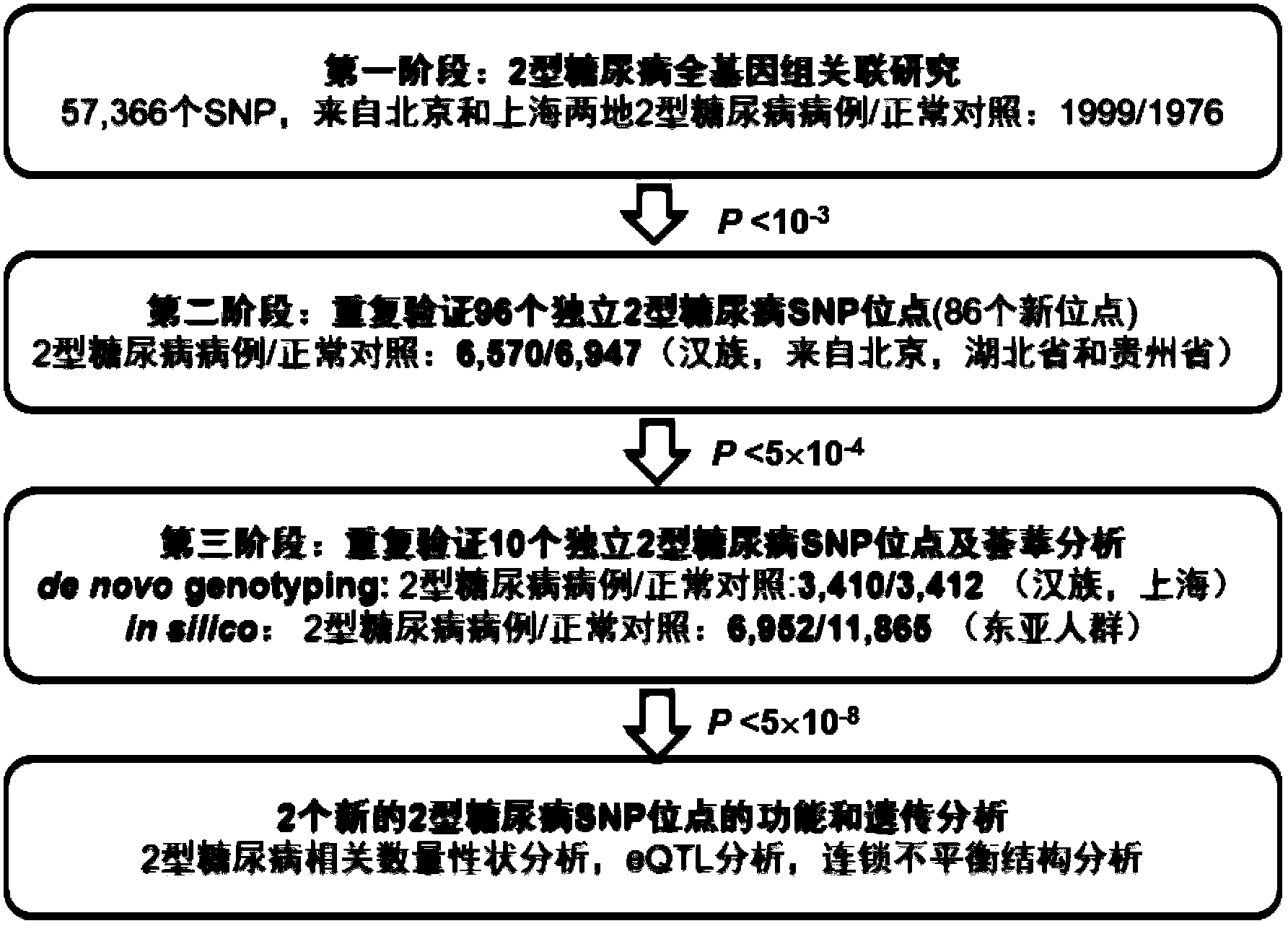
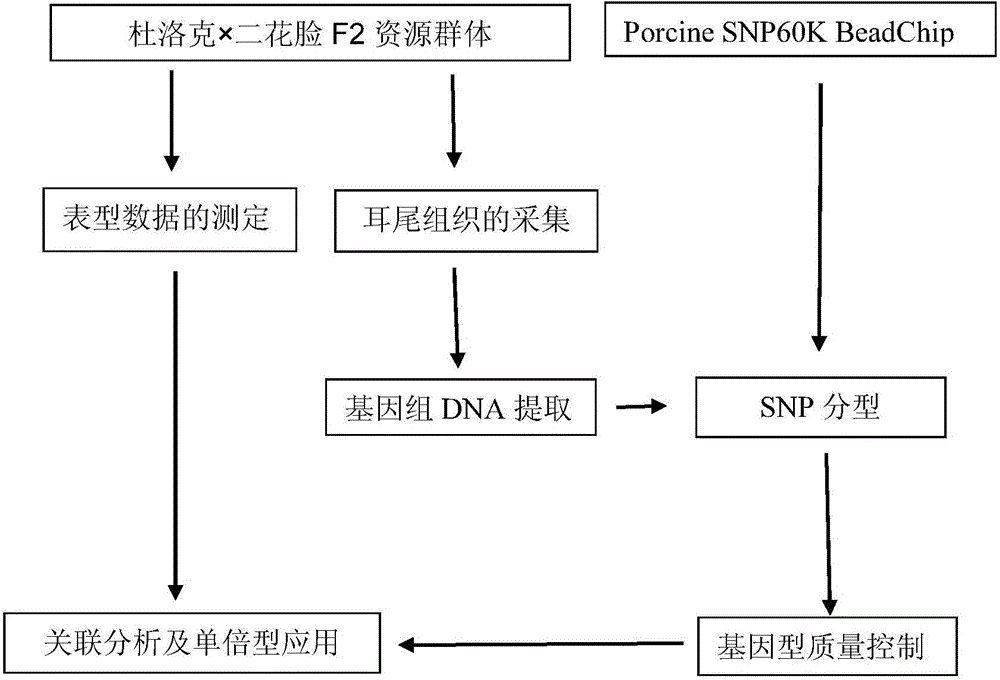
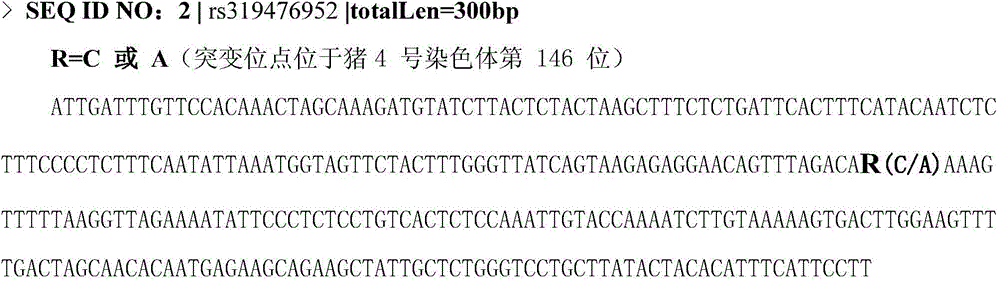
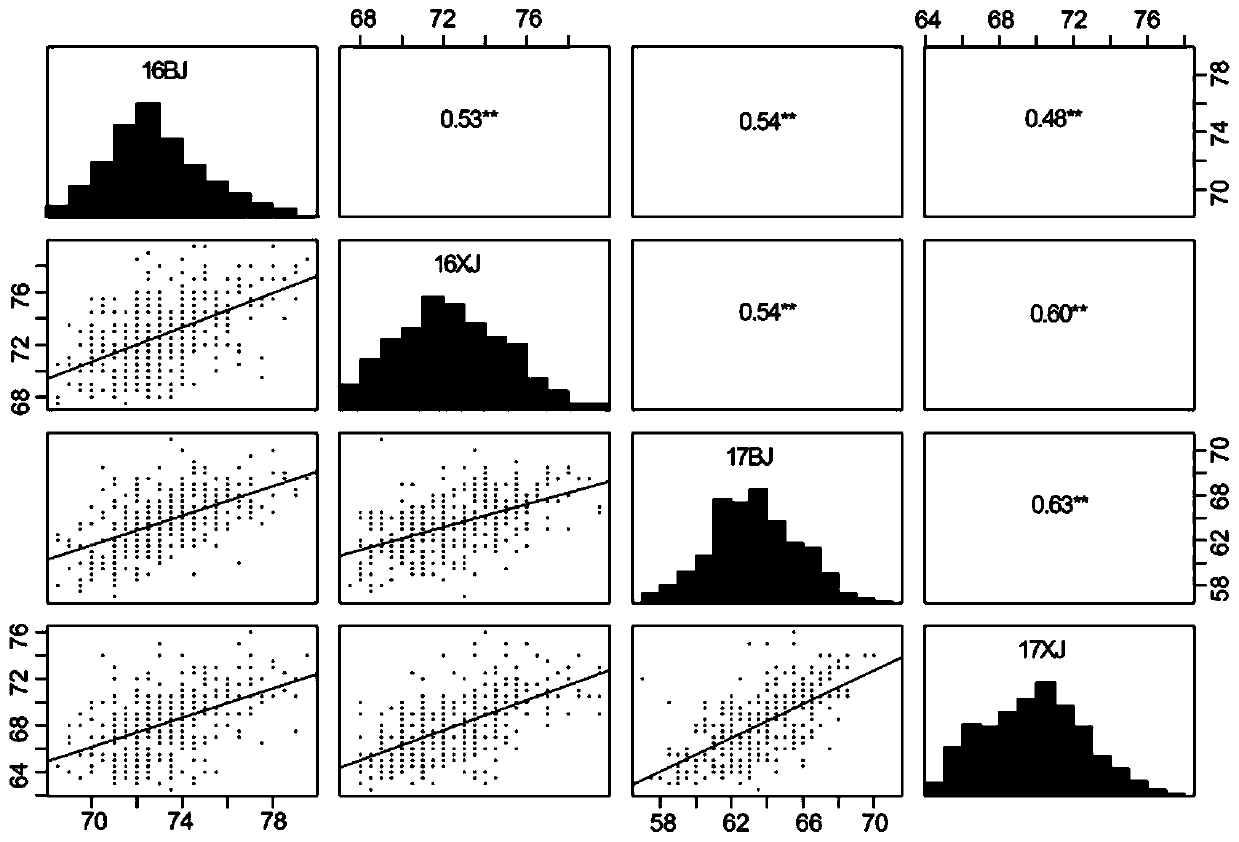
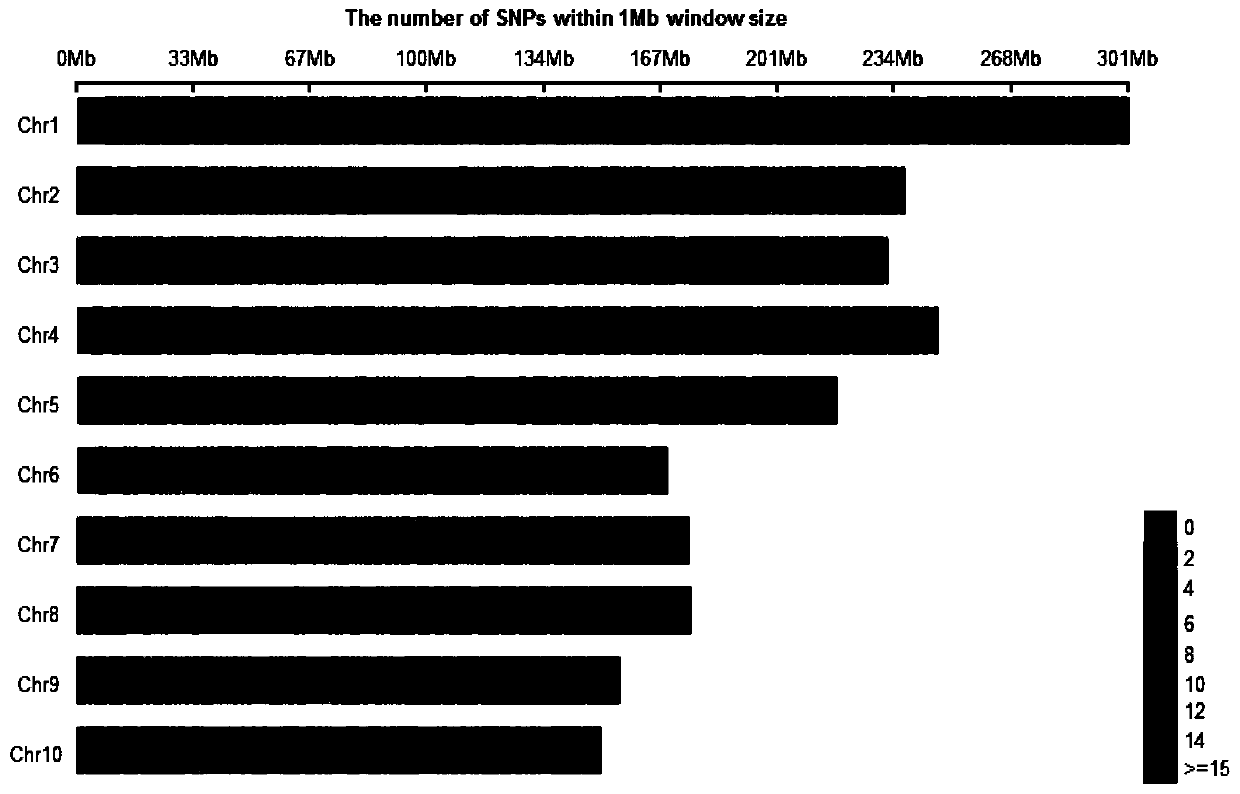
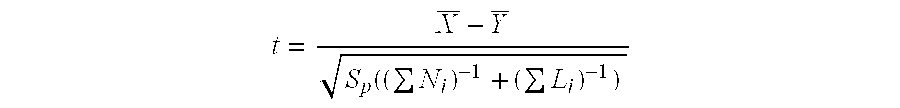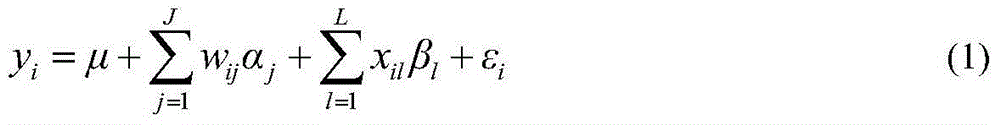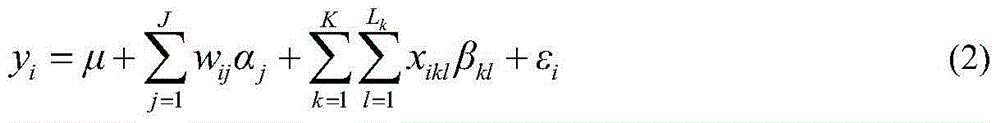Patents
Literature
62 results about "Genome-wide association study" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
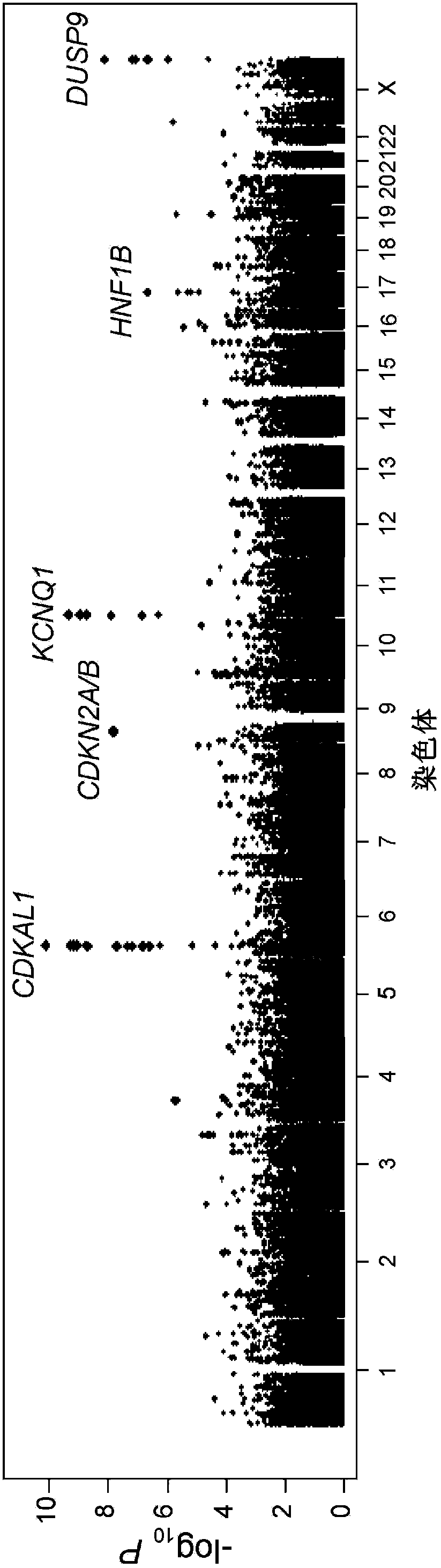
In genetics, a genome-wide association study (GWA study, or GWAS), also known as whole genome association study (WGA study, or WGAS), is an observational study of a genome-wide set of genetic variants in different individuals to see if any variant is associated with a trait. GWASs typically focus on associations between single-nucleotide polymorphisms (SNPs) and traits like major human diseases, but can equally be applied to any other genetic variants and any other organisms.
Genemap of the human genes associated with longevity
InactiveUS20090305900A1Electrolysis componentsVolume/mass flow measurementGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of SNP markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's longevity, their protection against age-related diseases and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
Genemap of the human genes associated with crohn's disease
InactiveUS20090081658A1Reduce frequencyTreatment safetyMicrobiological testing/measurementGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to Crohn's disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
Genetic diagnosis using multiple sequence variant analysis
The present invention is in the field of nucleic acid-based genetic analysis. More particularly, it discloses novel insights into the overall structure of genetic variation in all living species. The structure can be revealed with the use of any data set of genetic variants from a particular locus. The invention is useful to define the subset of variations that are most suited as genetic markers to search for correlations with certain phenotypic traits. Additionally, the insights are useful for the development of algorithms and computer programs that convert genotype data into the constituent haplotypes that are laborious and costly to derive in an experimental way. The invention is useful in areas such as (i) genome-wide association studies, (ii) clinical in vitro diagnosis, (iii) plant and animal breeding, (iv) the identification of micro-organisms.
Owner:METHEXIS GENOMICS
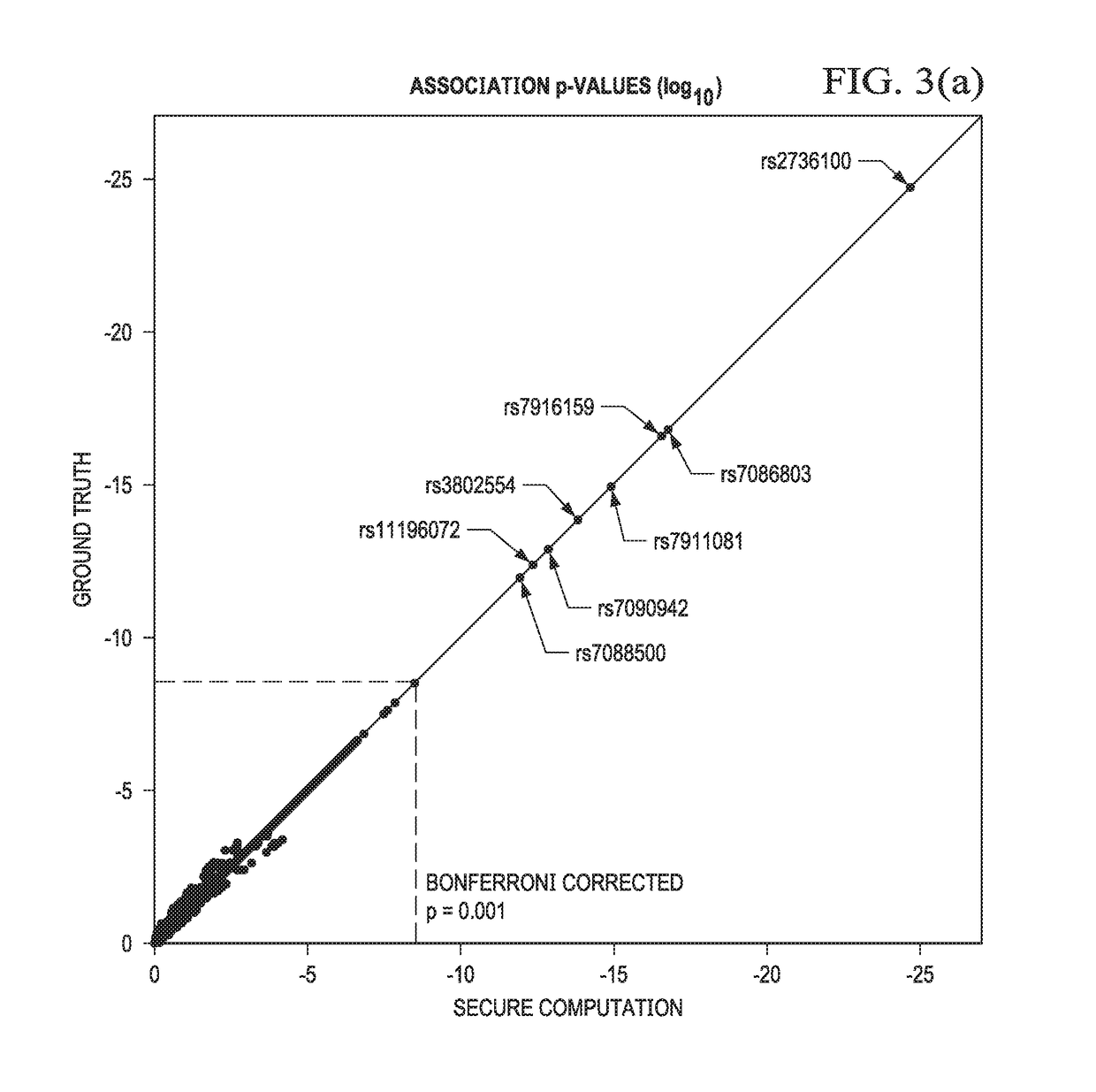
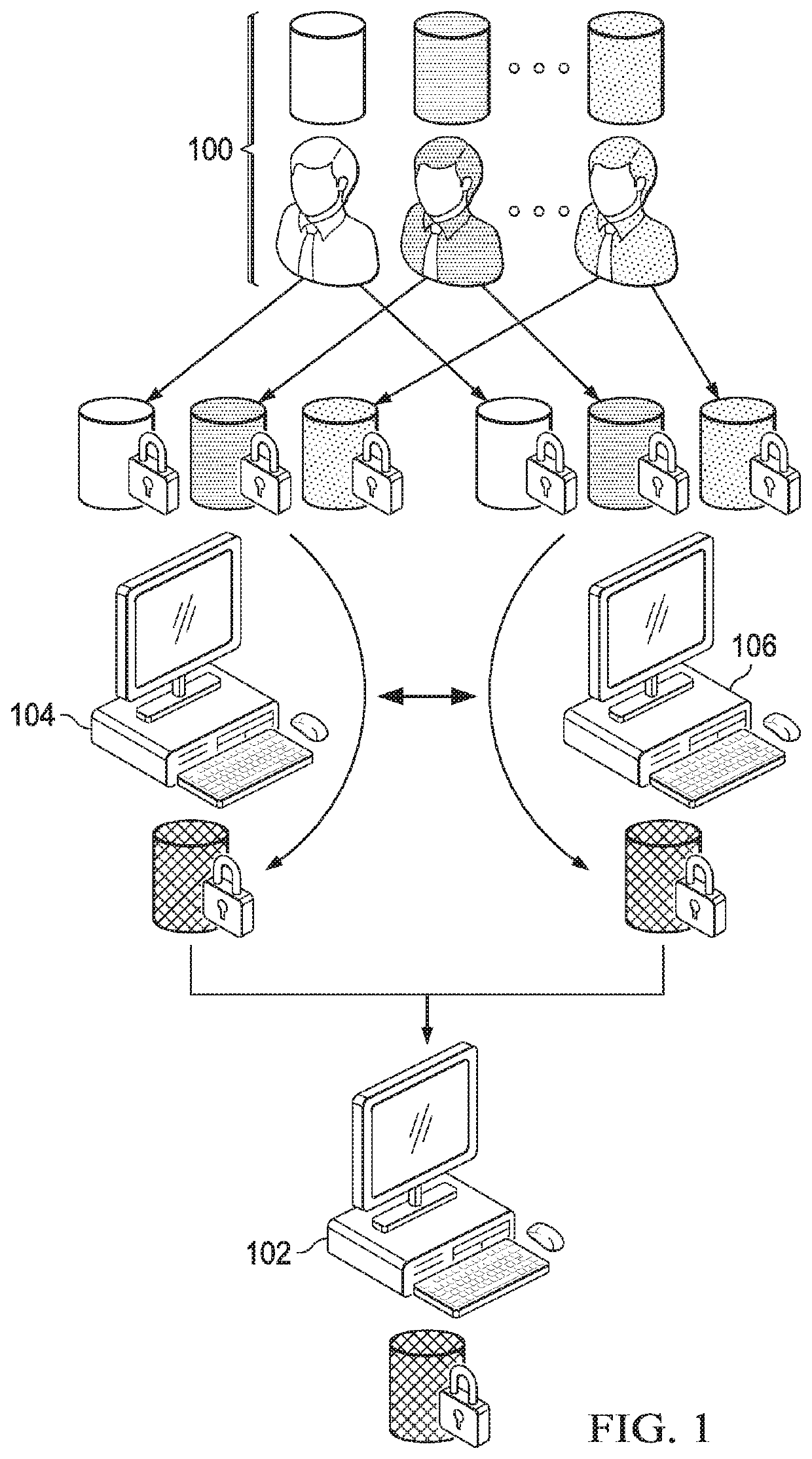
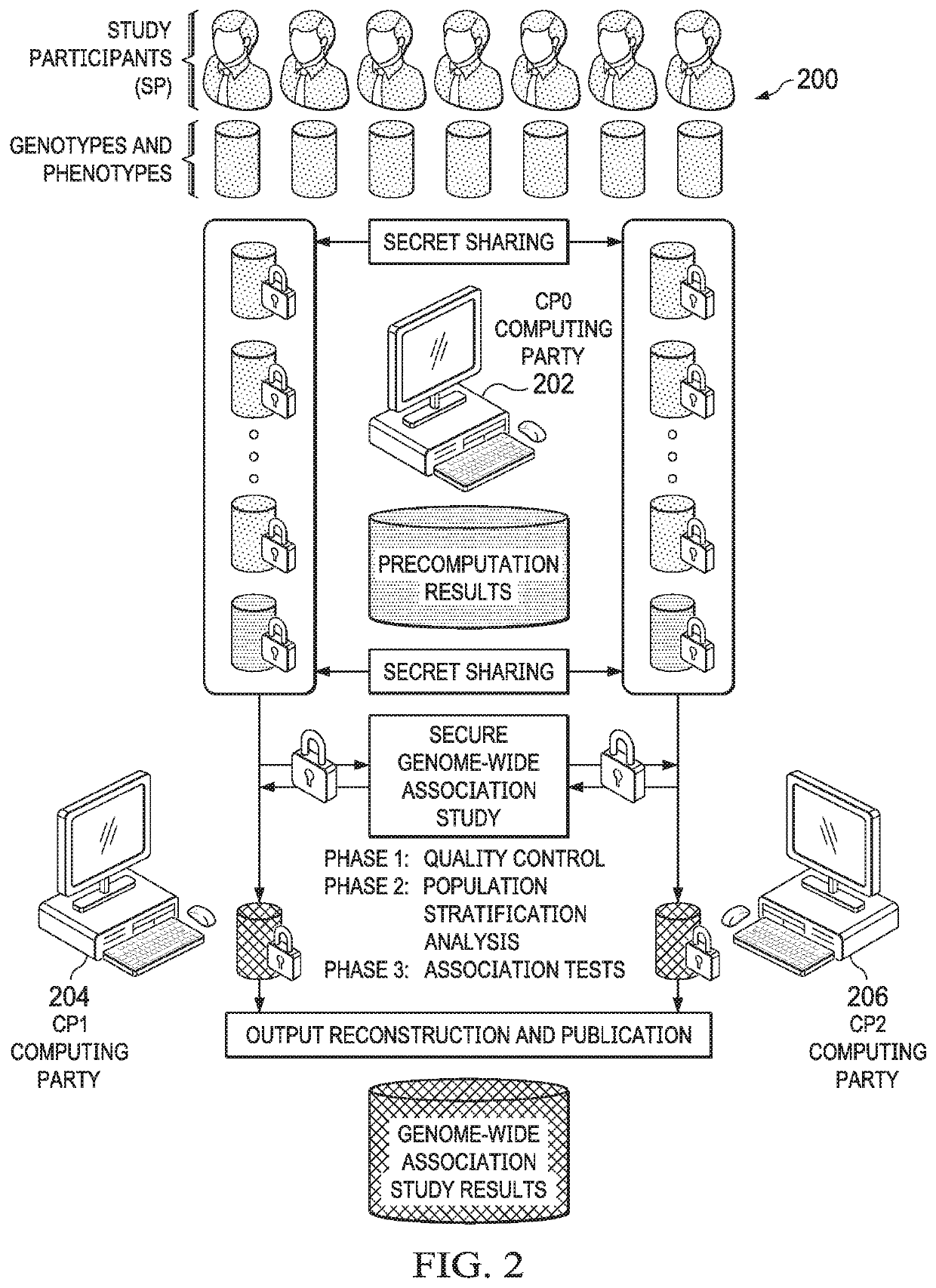
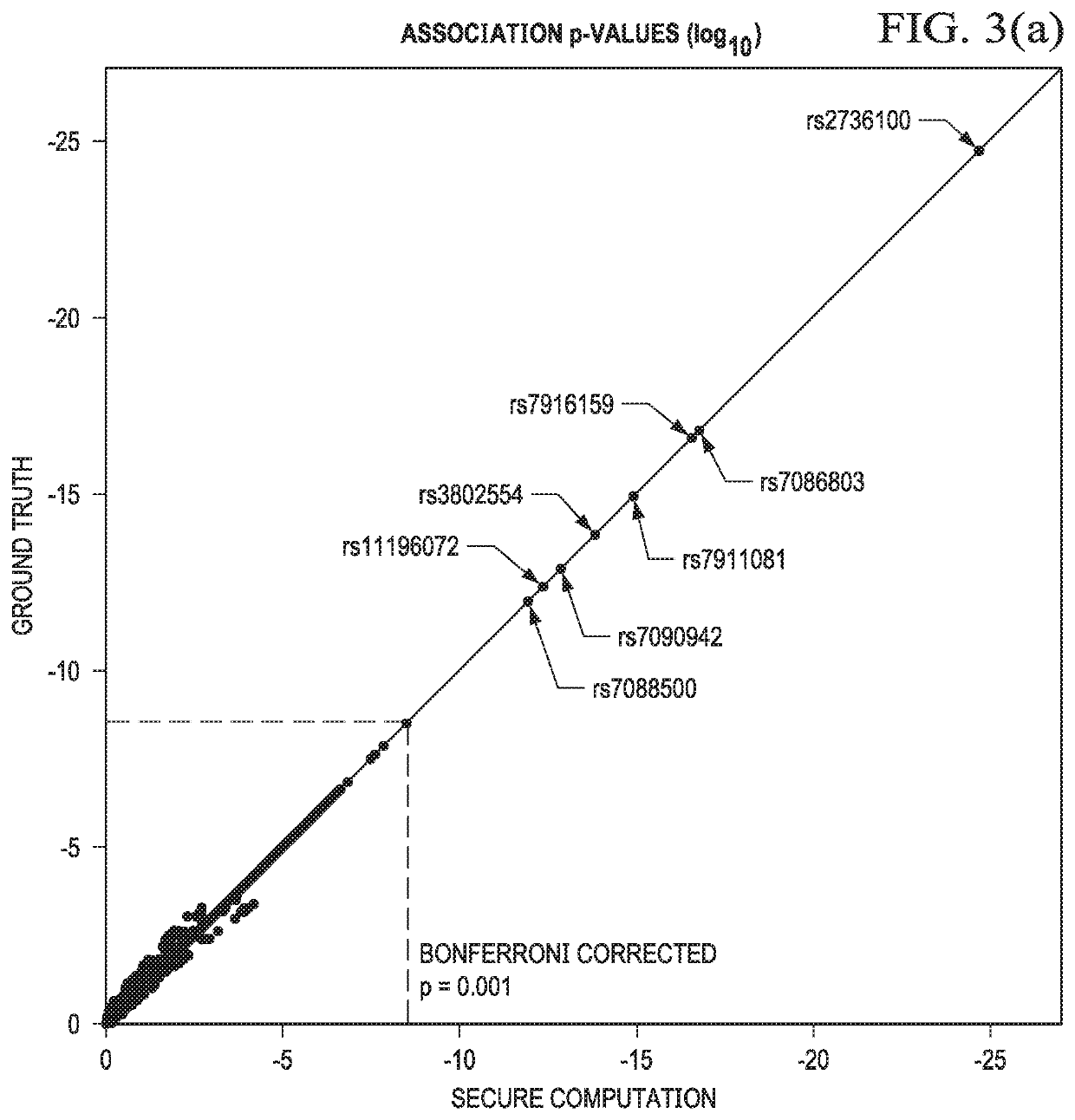
Secure genome crowdsourcing for large-scale association studies
ActiveUS20180373834A1Secure crowdsourcingPrivacy protectionProteomicsGenomicsData setIndividual study
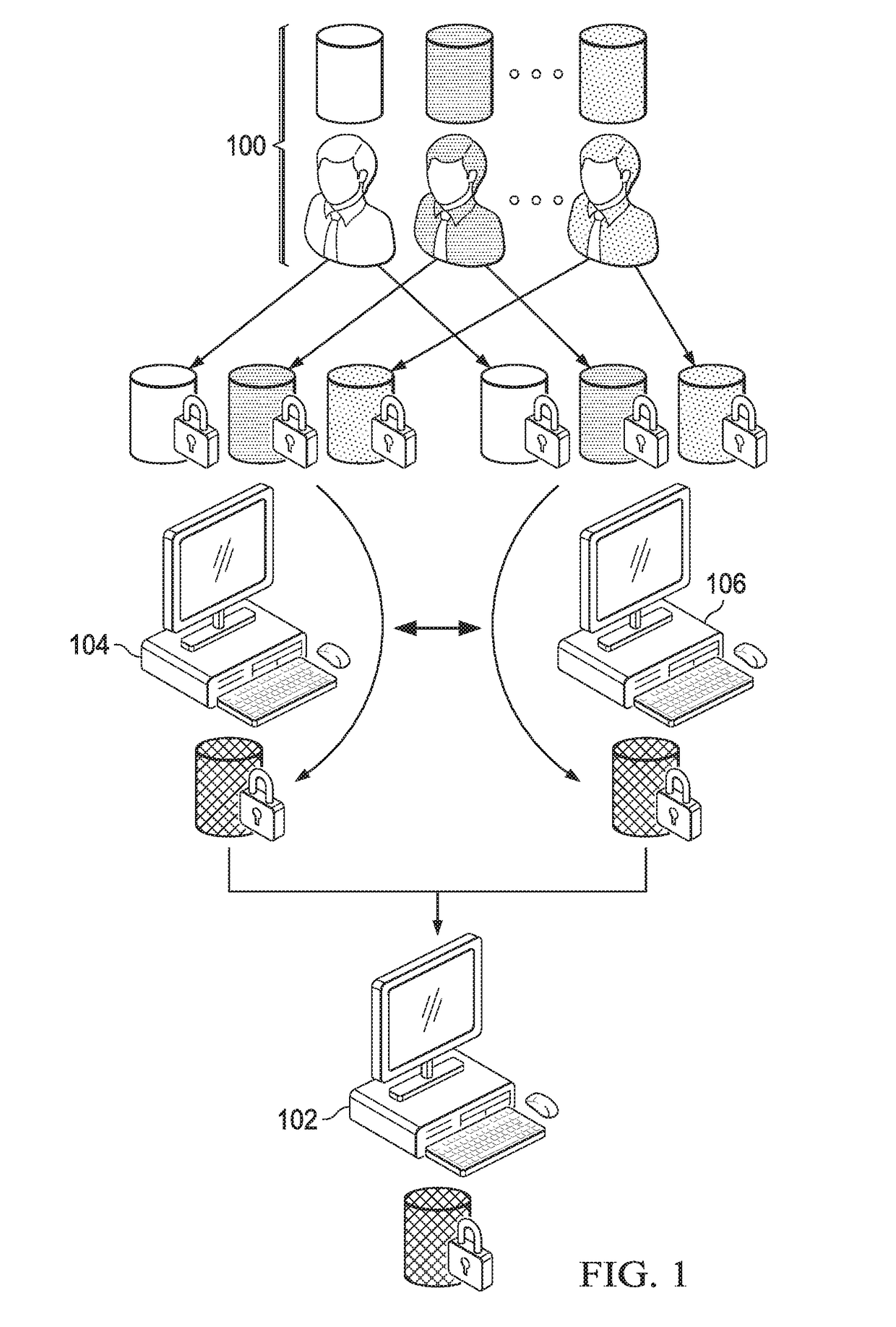
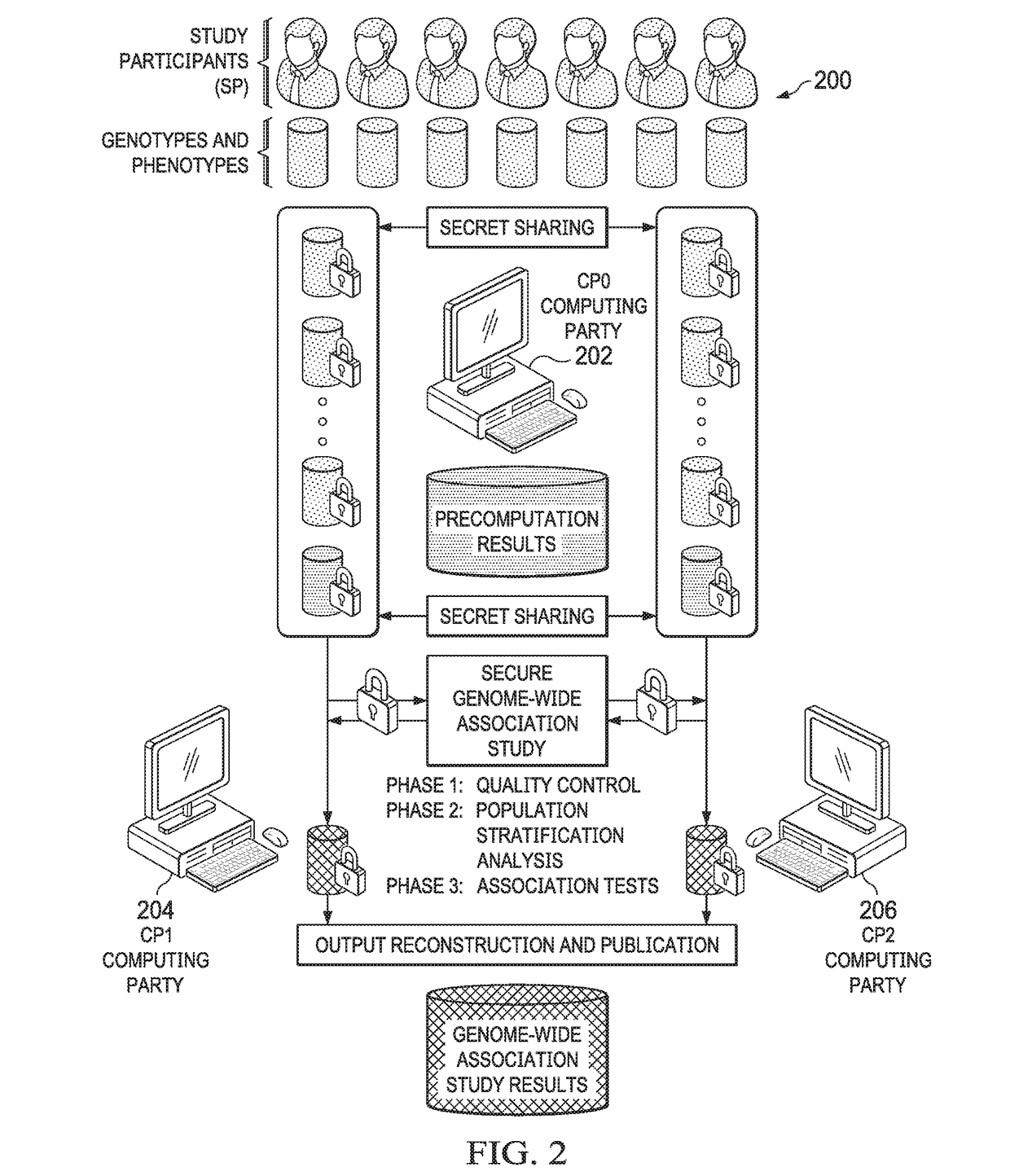
Computationally-efficient techniques facilitate secure crowdsourcing of genomic and phenotypic data, e.g., for large-scale association studies. In one embodiment, a method begins by receiving, via a secret sharing protocol, genomic and phenotypic data of individual study participants. Another data set, comprising results of pre-computation over random number data, e.g., mutually independent and uniformly-distributed random numbers and results of calculations over those random numbers, is also received via secret sharing. A secure computation then is executed against the secretly-shared genomic and phenotypic data, using the secretly-shared results of the pre-computation over random number data, to generate a set of genome-wide association study (GWAS) statistics. For increased computational efficiency, at least a part of the computation is executed over dimensionality-reduced genomic data. The resulting GWAS statistics are then used to identify genetic variants that are statistically-correlated with a phenotype of interest.
Owner:CHO HYUNGHOON +2
Genome-Wide Association Study Identifying Determinants Of Facial Characteristics For Facial Image Generation
InactiveUS20130039548A1Promote generationBiometric pattern for forensic purposeImage enhancementA-DNAOrganism
The present invention relates to a method for the generation of a facial composite from the genetic profile of a DNA-donor. The method comprises the steps of a) subjecting a biological sample to genotyping thereby generating a profile of genetic markers associated to numerical facial descriptors (NFD) for said sample, b) reverse engineer a NFD from the profile of the associated genetic variants and constructing a facial composite from the reverse engineered numerical facial descriptors (NFDs). The present invention also relates to a method for identifying genetic markers and / or combinations of genetic markers that are predictive of the facial characteristics, (predictive facial markers) of a person, said method comprising the steps of: a) capturing images of a group of individual faces; b) performing image analysis on facial images of said group of individual faces thereby extracting phenotypical descriptors of the faces; c) obtaining data on genetic variation from said group of individual and d) performing a genome-wide association study (GWAS) to identify said predictive facial markers.
Owner:DANMARKS TEKNISKE UNIV
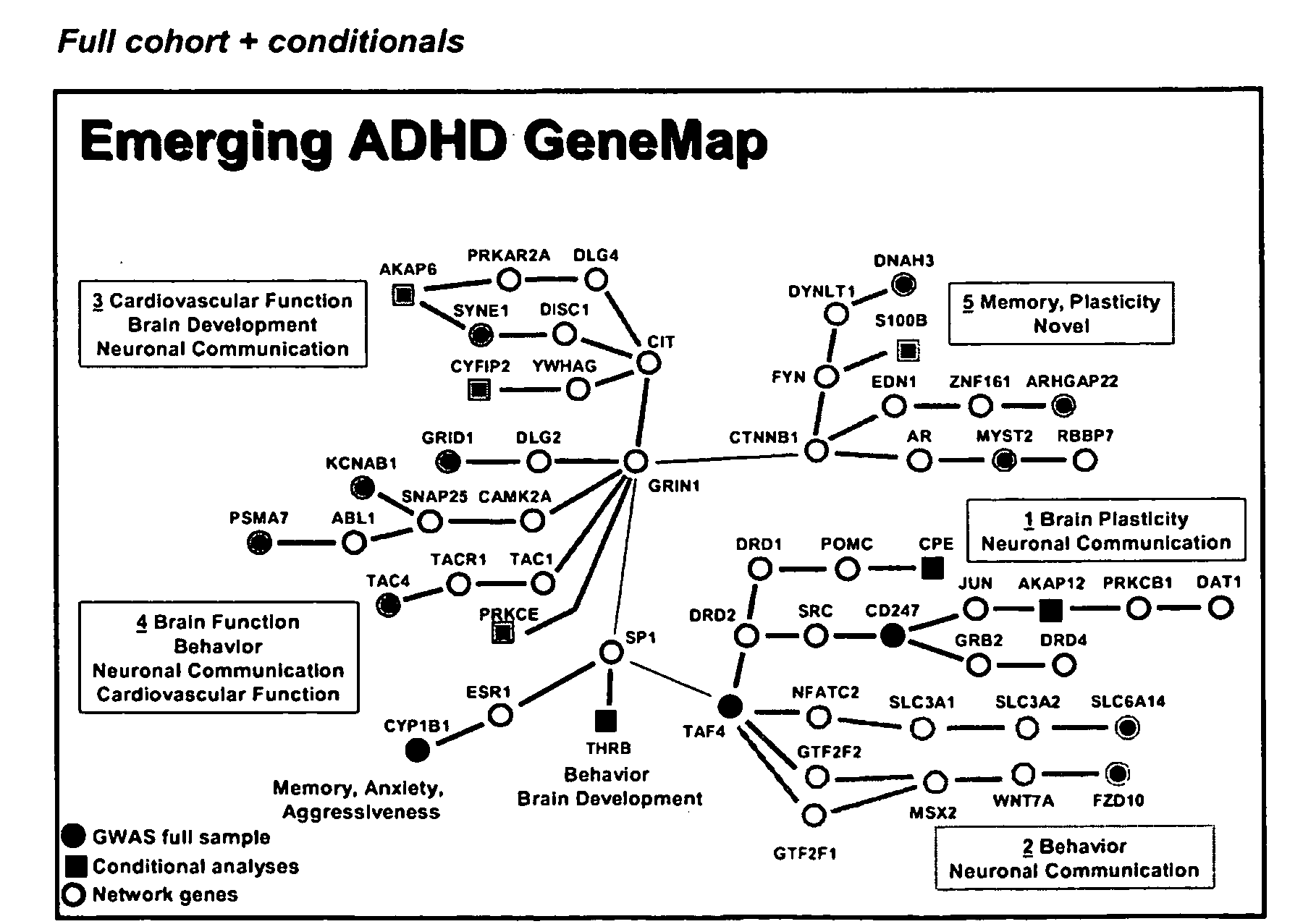
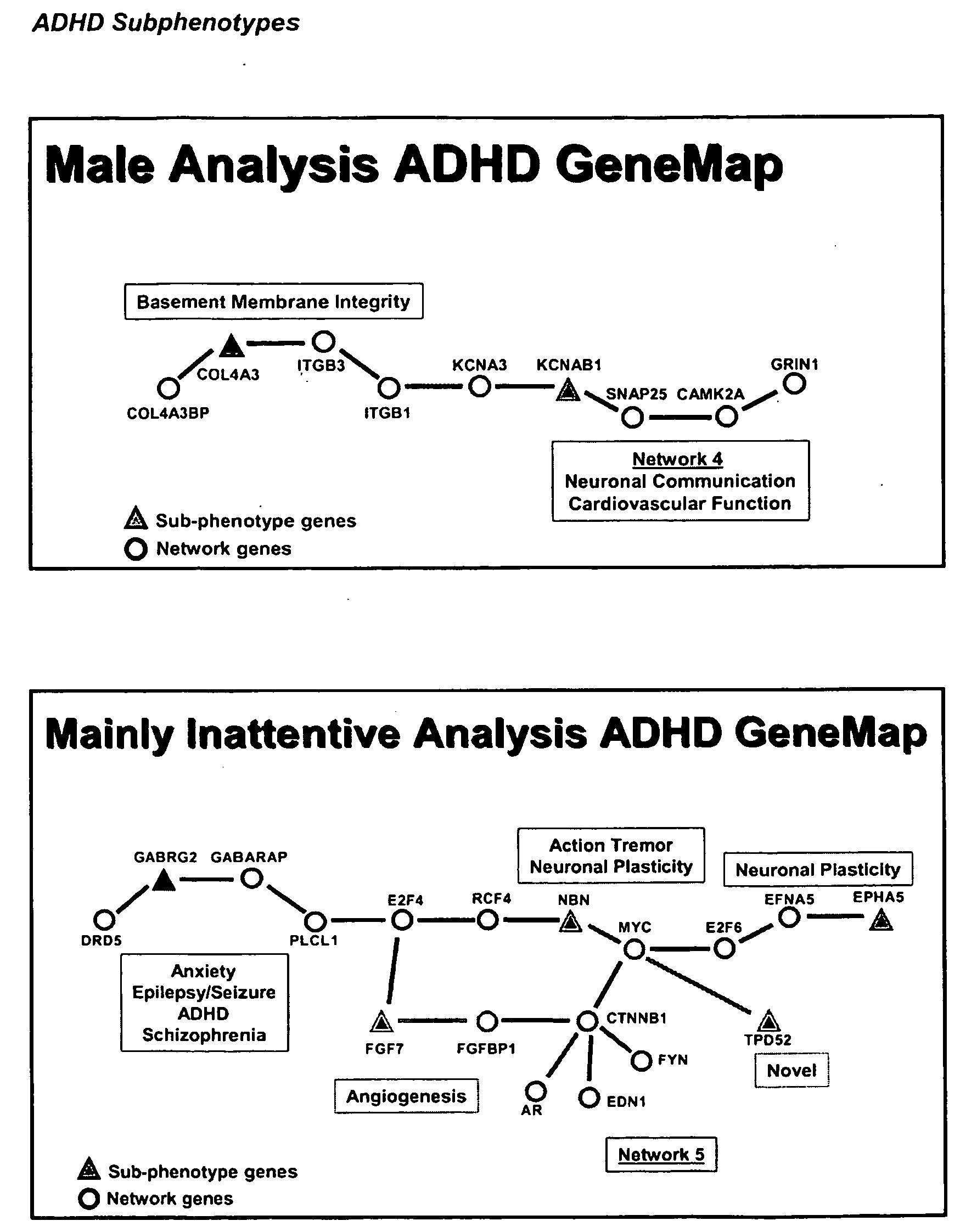
Genemap of the human genes associated with adhd
InactiveUS20100120628A1Easy to understandConvenient treatmentMicrobiological testing/measurementLibrary screeningGenomicsDisease
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to ADHD disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
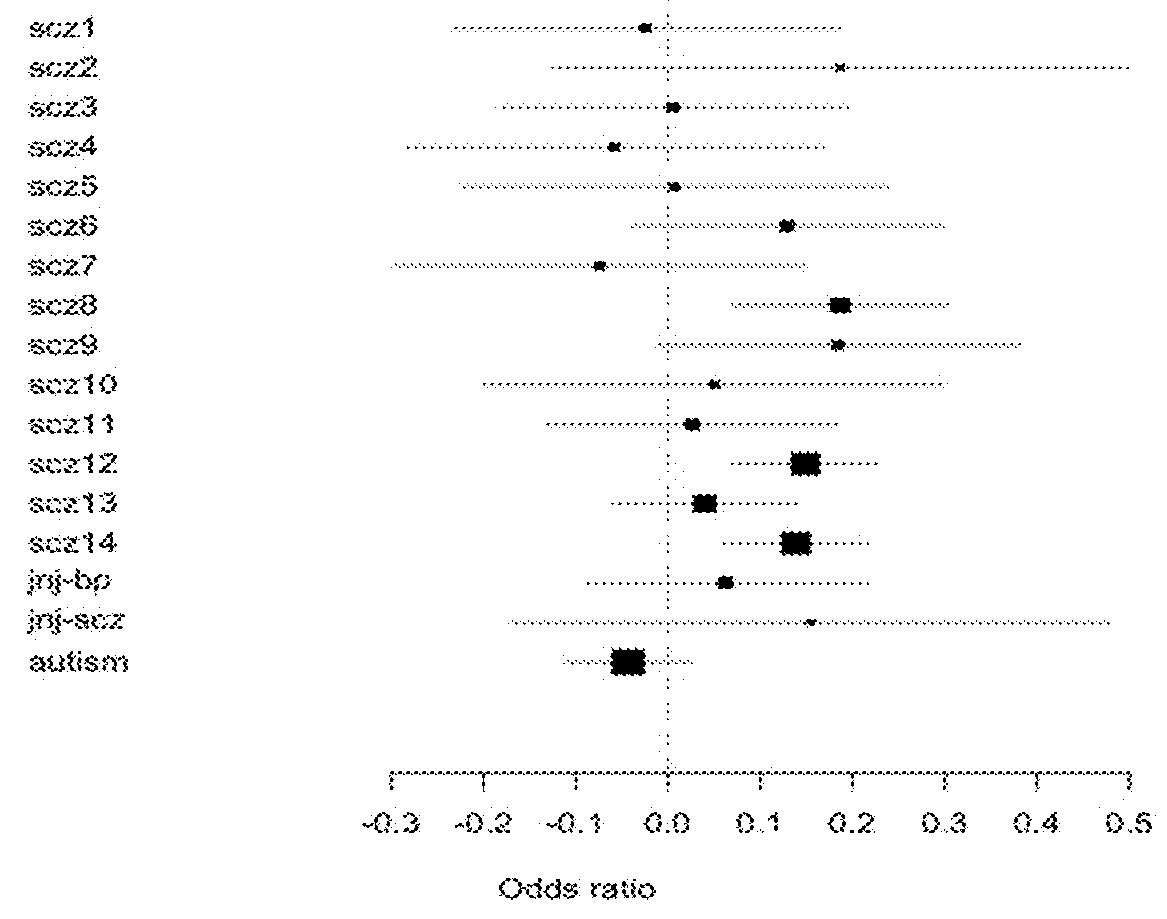
Schizophrenia-associated genetic loci identified in genome wide association studies and methods of use thereof
Compositions and methods for the identification of agents useful for the treatment of neurological disorders, including schizophrenia, are provided.
Owner:THE CHILDRENS HOSPITAL OF PHILADELPHIA
Genemap of the human genes associated with crohn's disease
InactiveUS20090181380A1Microbiological testing/measurementAnalogue computers for chemical processesGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism makers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to IBD (ex: Chrohn's disease) and / or their response to a particular drug or drugs.
Owner:BELOUCHI ABDELMAJID +12
SNP (Single Nucleotide Polymorphism) marker related to bitter character of cucumber and application of SNP marker
ActiveCN103013966ASpeed up the breeding processEasy to identifyFungiBacteriaPhenotypic traitBitter taste
The invention provides an SNP (Single Nucleotide Polymorphism) marker related to the bitter character of cucumber and application of the SNP marker. The SNP marker is located at the NO.1178bp of the gene sequence for coding the Csa6G088690 protein of cucumber; cucumber with basic groups G at the NO.1178bp is bitter; and cucumber with basic groups A at the NO.1178bp is not bitter. According to the invention, a genome wide association study (GWAS) method is adopted to screen out the SNP marker related to the bitter taste character of cucumber, a derived cleaved amplified polymorphic sequences (dCAPS) method is adopted to detect the gene types of cucumber to be detected, and bitter or non-bitter cucumber variety breeding is carried out according to the gene types, so as to speed up the breeding process of non-bitter cucumber. The SNP marker is more accurate and reliable in result than the traditional method of subjectively judging whether cucumber is bitter or not, is simpler and more convenient than HPLC (High Performance Liquid Chromatography) for identifying bitter substances, and can be used for large-scale screening of breeding materials. Besides, the invention discovers the committed step for the Csa6G088690 protein of cucumber to control the synthesis pathway of bitter of cucumber for the first time.
Owner:INST OF VEGETABLE & FLOWERS CHINESE ACAD OF AGRI SCI
Genemap of the human genes associated with crohn's disease
InactiveUS20100081129A1Genetic material ingredientsMicrobiological testing/measurementLinkage Disequilibrium MappingPharmacogenomic Study
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to Crohn's disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
Genemap of the human genes associated with schizophrenia
InactiveUS20100144538A1Immobilised enzymesBioreactor/fermenter combinationsGenomicsLinkage Disequilibrium Mapping
The present invention relates to the selection of a set of polymorphism markers for use in genome wide association studies based on linkage disequilibrium mapping. In particular, the invention relates to the fields of pharmacogenomics, diagnostics, patient therapy and the use of genetic haplotype information to predict an individual's susceptibility to SCHIZOPHRENIA disease and / or their response to a particular drug or drugs.
Owner:GENIZON BIOSCI
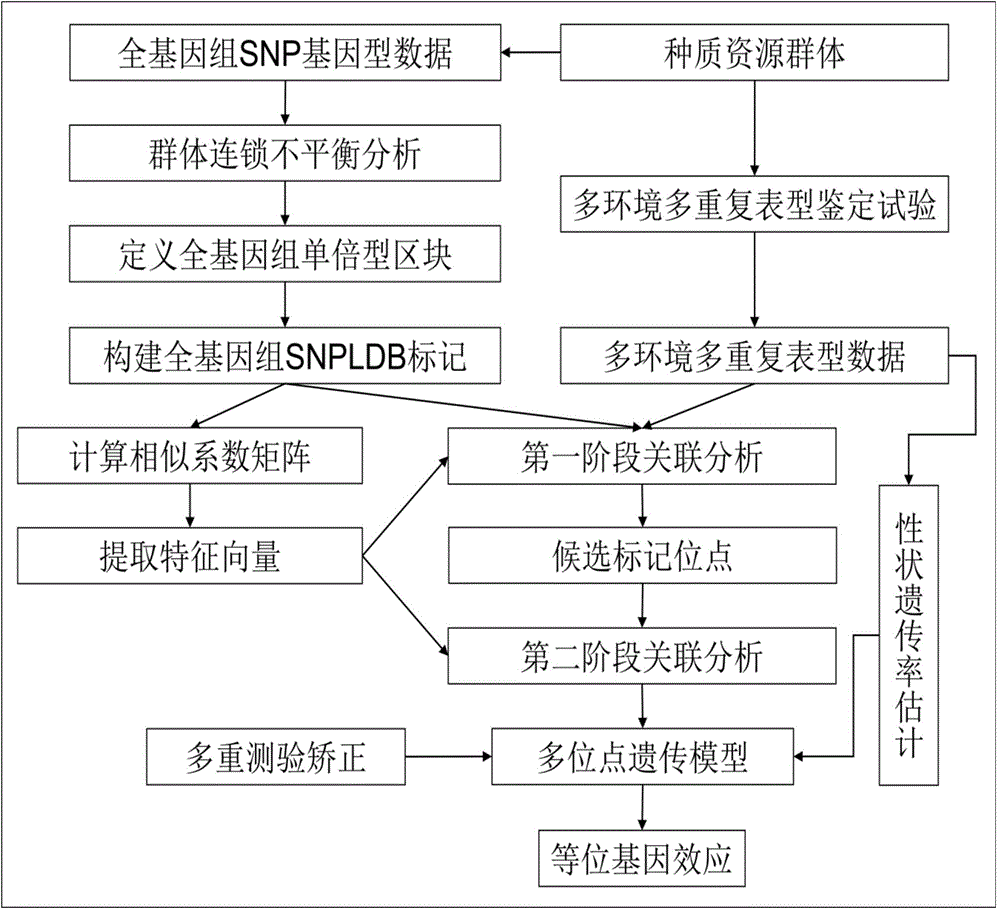
Restrictive two-stage genome-wide association study (GWAS) method based on SNPLDB mark
InactiveCN104651517AShorten the attenuation distanceMultiple allelic variationMicrobiological testing/measurementProteomicsMulti siteRegression analysis
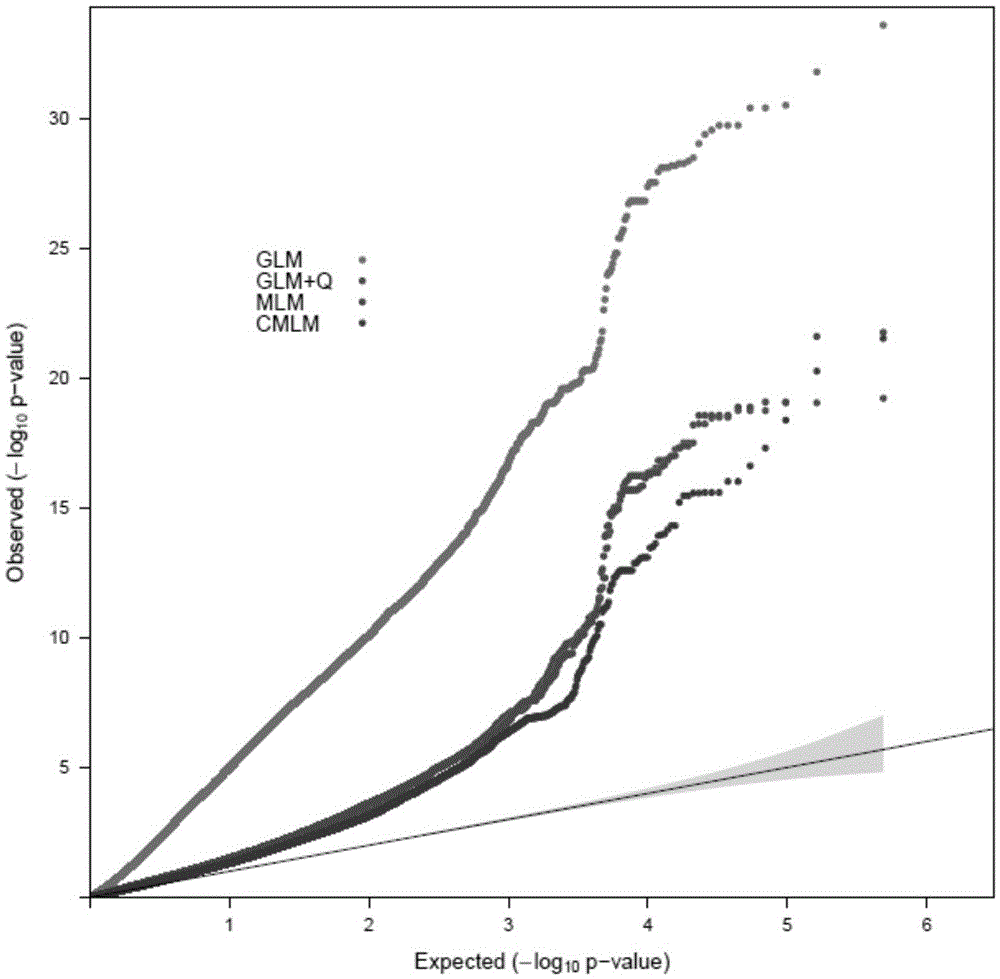
The invention discloses a restrictive two-stage genome-wide association study (GWAS) method based on an SNPLDB mark, and aims at solving the problems that a traditional method cannot be used for estimating multiple allele information, and is high in false positive rate and low in efficiency of detection on inbreeding crops. By combination of the SNPLDB mark constructed on the basis of a haplotype block, correction of deviation of an inbreeding population relation analysis model and a two-stage relation analysis strategy under a multi-site model, the GWAS method suitable for conventional breeding of inbreeding crops is built; the SNPLDB mark is applied to GWAS, so that a method is provided for multiple allele estimation; candidate sites are screened on the basis of a single-site model in the first stage, and are further screened on the basis of a progressive regression analysis method in the multi-site model in the second stage, so as to balance the problems of missing of heritability and over-high heritability estimation. Therefore, the interpretation ratio of a final genetic model is controlled at trait heritability. The positioning accuracy and efficiency are improved by a feature vector and an appropriate significant level of a similarity coefficient matrix estimated by the SNPLDB mark by GWAS.
Owner:NANJING AGRICULTURAL UNIVERSITY
Colorectal cancer susceptibility diagnostic kit and application of SNP (single nucleotide polymorphism) in preparation of diagnostic kit
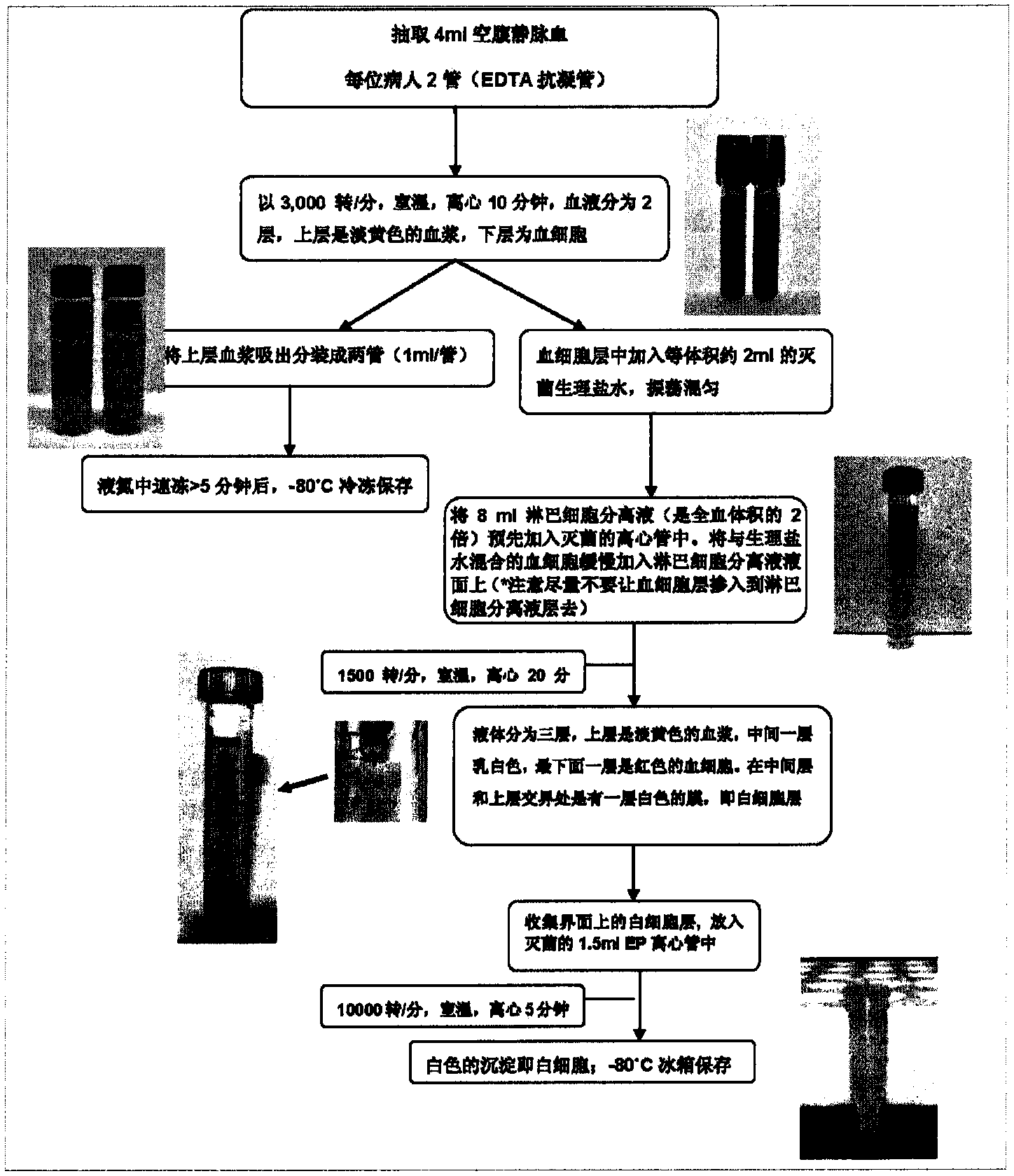
The invention discloses a colorectal cancer susceptibility diagnostic kit and an application of SNP (single nucleotide polymorphism) in preparation of the diagnostic kit. The SNP sites are rs12522693 and rs17836917. By means of genome-wide association study, the inventors prove remarkable correlation between an allele at the rs12522693 site G and an allele at the rs17836917 site A and colorectal cancer susceptibility through a screening stage, first stage verification and second stage verification. By means of detecting the genotypes of the rs12522693 site and / or the rs17836917 site, the colorectal cancer susceptibility of a testee can be predicted, and thus early prevention and treatment of the disease can be promoted. The colorectal cancer susceptibility diagnostic kit can be used for detecting the genotypes of the rs12522693 site and / or the rs17836917 site by using known technologies in the field so as to judge whether the testee has colorectal cancer susceptibility or not.
Owner:PEOPLES HOSPITAL PEKING UNIV
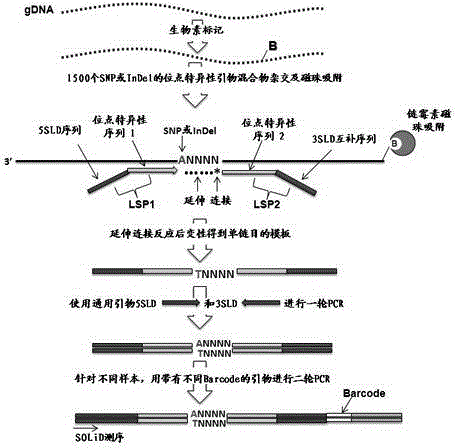
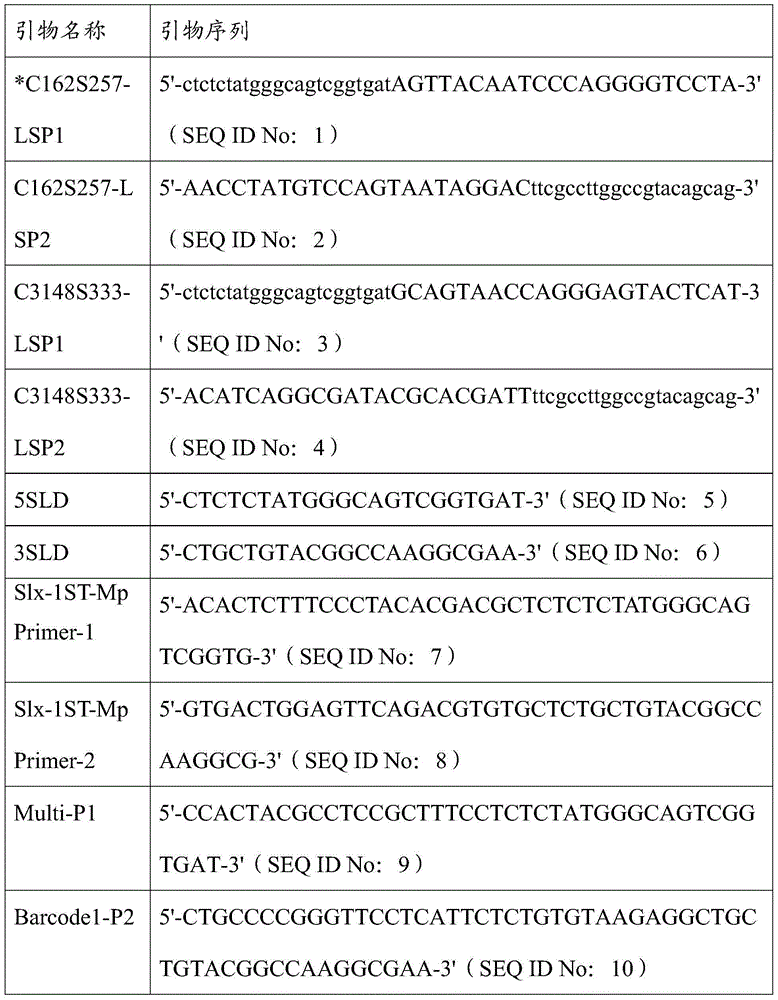
High-throughput low-cost SNP (single nucleotide polymorphism) genotyping method based on liquid molecular hybridization principle
ActiveCN103555846AImprove throughputImprove accuracyMicrobiological testing/measurementPhosphorylationBiotin
The invention provides a high-throughput low-cost SNP (single nucleotide polymorphism) genotyping method based on a liquid molecular hybridization principle. The method comprises the following steps: extracting biological genome DNA (deoxyribonucleic acid) and carrying out biotin labeling on the biological genome DNA; designing site-specific hybridization primers LSP1 and LSP2 and carrying out 5' phosphorylation on LSP2; hybridizing an LSP1 mixture and an LSP2 mixture with the genome DNA to obtain a hybridization adsorption product; carrying out extended linkage reaction to obtain a target DNA fragment; carrying out a round of PCR (polymerase chain reaction) amplification on a universal primer; carrying out PCR amplification on the Barcode specific primer of the recovered target fragment; carrying out high-throughput sequencing; obtaining SNP site genotyping information through analysis. The method combines the site selection flexibility of the liquid hybridization technology with the advantages of high throughput and low cost of the high-throughput sequencing technology and has great application value and wide popularization prospect in the research fields such as large-scale screening of SNP, genome-wide association study, population diversity evaluation, gene function analysis and the like.
Owner:OCEAN UNIV OF CHINA
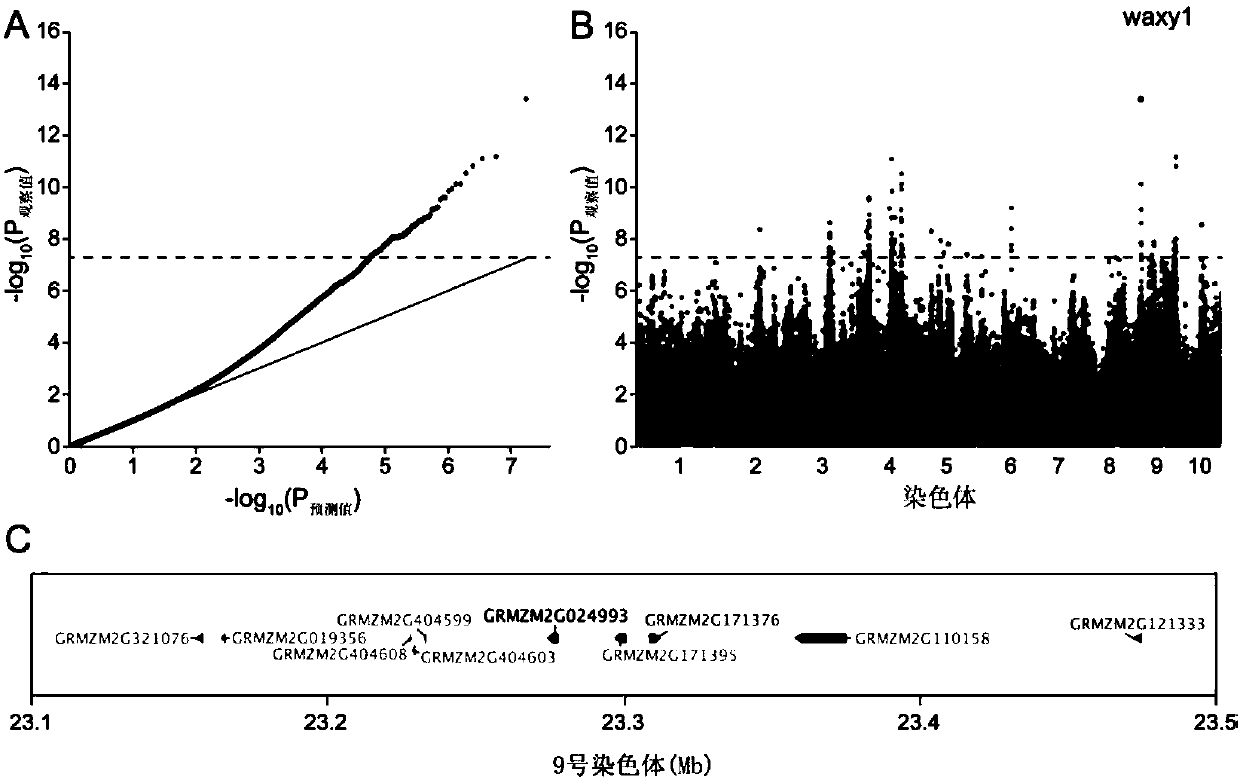
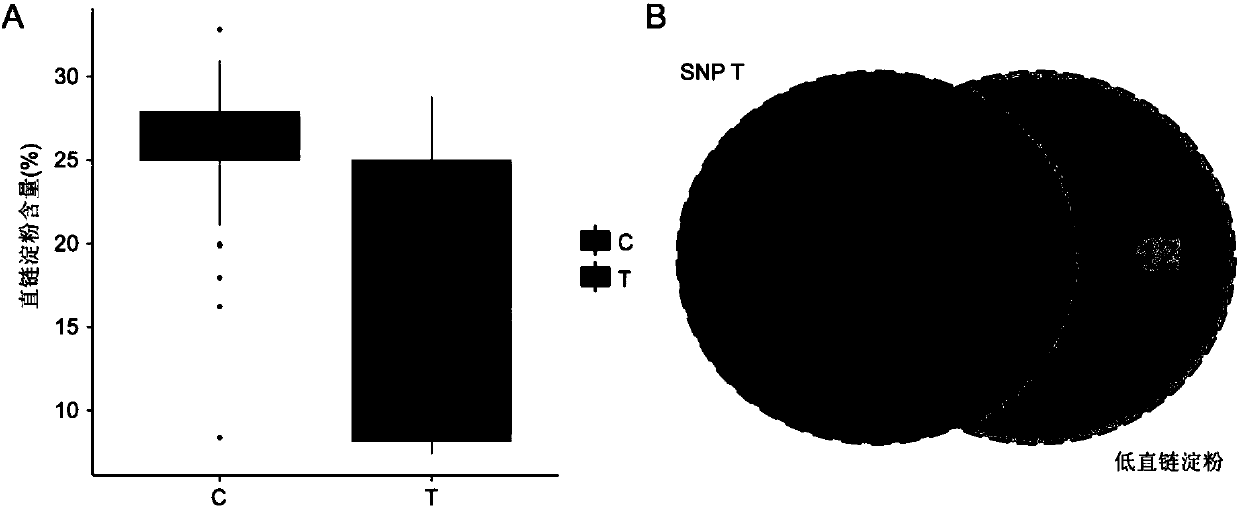
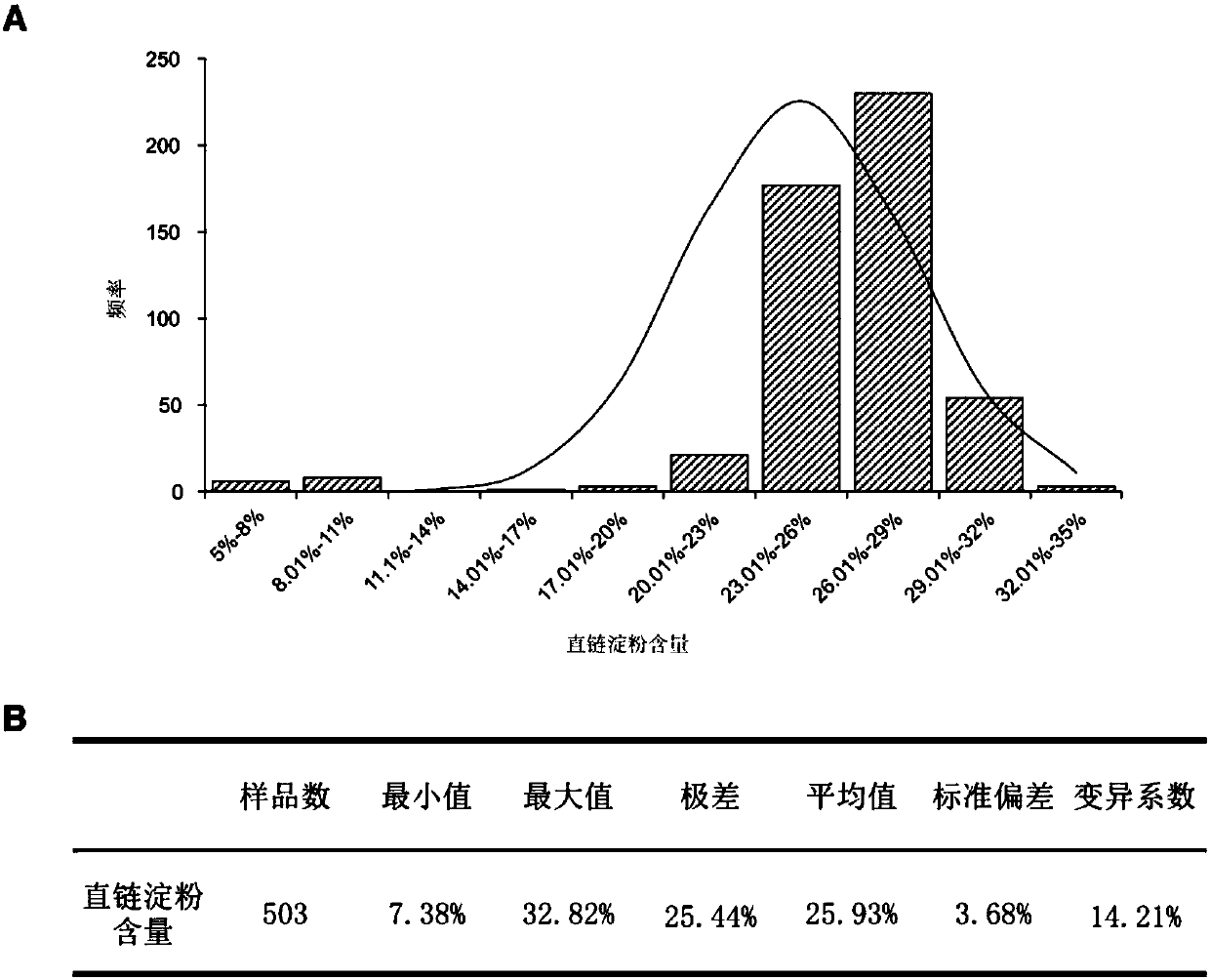
Method for developing waxy1-gene internal molecular markers on basis of association analysis and KASP
ActiveCN107619873AQuick digRapid phenotypeMicrobiological testing/measurementDNA/RNA fragmentationAmylaseCandidate Gene Association Study
The invention discloses a method for developing waxy1-gene internal molecule markers on the basis of association analysis and KASP. One indel and two snap markers located in waxy1 gene and remarkablyassociated with content of amylase are found out mainly through genome-wide association study and candidate gene association analysis techniques; the detected indel is marked as a major mutant type ofglutinous maize in China; glutinous maize materials can be identified through agarose gel by the markers developed on the basis of the indel, and convenience and rapidness is achieved as compared with that of original polyacrylamide gel identification; meanwhile, paired linkage disequilibrium test of 59 parts of waxy1-gene sequence regions of maize hybrid line materials is performed, close linkage of the indel marker and the two snp markers is tested, primers of the two snps are developed on the basis of the KASP technology, the glutinous maize materials can be identified rapidly, efficientlyand accurately, and molecular breeding of glutinous maize is facilitated.
Owner:SHANGHAI JIAO TONG UNIV
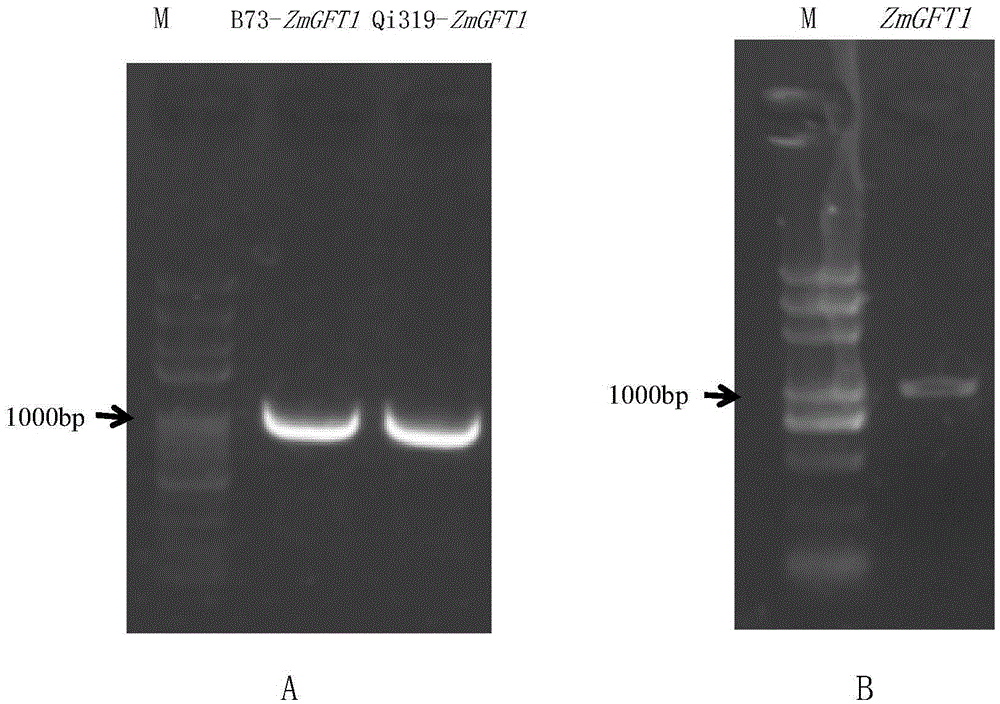
Application of maize gene ZmGFT1 in increasing folic acid content in plants
ActiveCN105647942AAlleviate hidden hungerMicrobiological testing/measurementTransferasesBiotechnologyNutrition
The invention provides application of maize gene ZmGFT1 in increasing folic acid content in plants. 5-Formyltetrahydrofolate content in 513 selfing line materials of a maize natural variant group is measured, the influence of group structure and genetic relationship of the natural variant group is estimated in connection with genotype data mined from RNA-Seq data, gene GFT1 having significant influence on the 5-formyltetrahydrofolate content and located on maize 5th chromosome is located by means of genome-wide association study, and the gene GFT1 encodes glutamate formiminotransferase 1. The invention also provides an SNP marker associated with maize folic acid content and its application. The maize gene ZmGFT1 and discovery of the key SNP site therein having significant influence on the 5-formyltetrahydrofolate content not only provide theoretical basis for studying folic acid metabolism in maize and other crops, but also enable the hidden hunger problem of human in terms of folic acid nutrition to be solved through the increase in the folic acid content of transgenic plants.
Owner:THE INST OF BIOTECHNOLOGY OF THE CHINESE ACAD OF AGRI SCI
Genetic diagnosis using multiple sequence variant analysis
The present invention is in the field of nucleic acid-based genetic analysis. More particularly, it discloses novel insights into the overall structure of genetic variation in all living species. The structure can be revealed with the use of any data set of genetic variants from a particular locus. The invention is useful to define the subset of variations that are most suited as genetic markers to search for correlations with certain phenotypic traits. Additionally, the insights are useful for the development of algorithms and computer programs that convert genotype data into the constituent haplotypes that are laborious and costly to derive in an experimental way. The invention is useful in areas such as (i) genome-wide association studies, (ii) clinical in vitro diagnosis, (iii) plant and animal breeding, (iv) the identification of micro-organisms.
Owner:METHEXIS GENOMICS
Secure secret-sharing-based crowdsourcing for large-scale association studies of genomic and phenotypic data
ActiveUS10910087B2Facilitate secure crowdsourcingPrivacy protectionProteomicsGenomicsData setGenomic data
Owner:CHO HYUNGHOON +2
Method for discovering pharmacogenomic biomarkers
InactiveUS20140031242A1Quality improvementPredict likelihood of successSugar derivativesMicrobiological testing/measurementPharmacogenomicsDrugs response
The present invention relates to a method of discovering pharmacogenomic biomarkers that are correlated with varied individual responses (efficacy, adverse effect, and other end points) to therapeutic agents. The present invention provides a mean to utilize archived clinical samples to perform genome-wide association study in order to identify novel pharmacogenomic biomarkers. The newly discovered biomarkers can then be developed into companion diagnostic tests which can help to predict drug responses and apply drugs only to those who will be benefited, or exclude those who might have adverse effects, by the treatment.
Owner:DENOVO BIOPHARMA HANGZHOU LTD
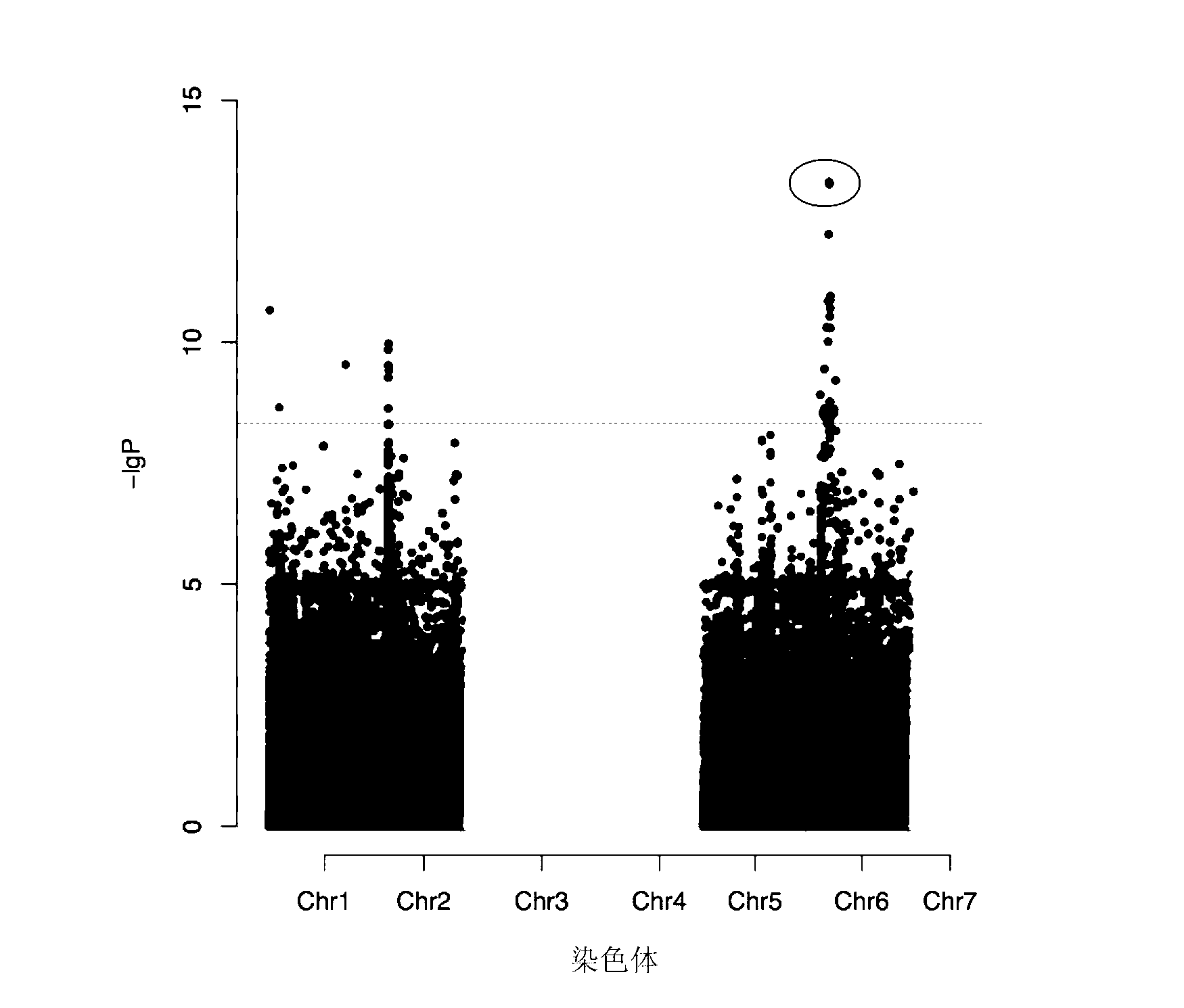
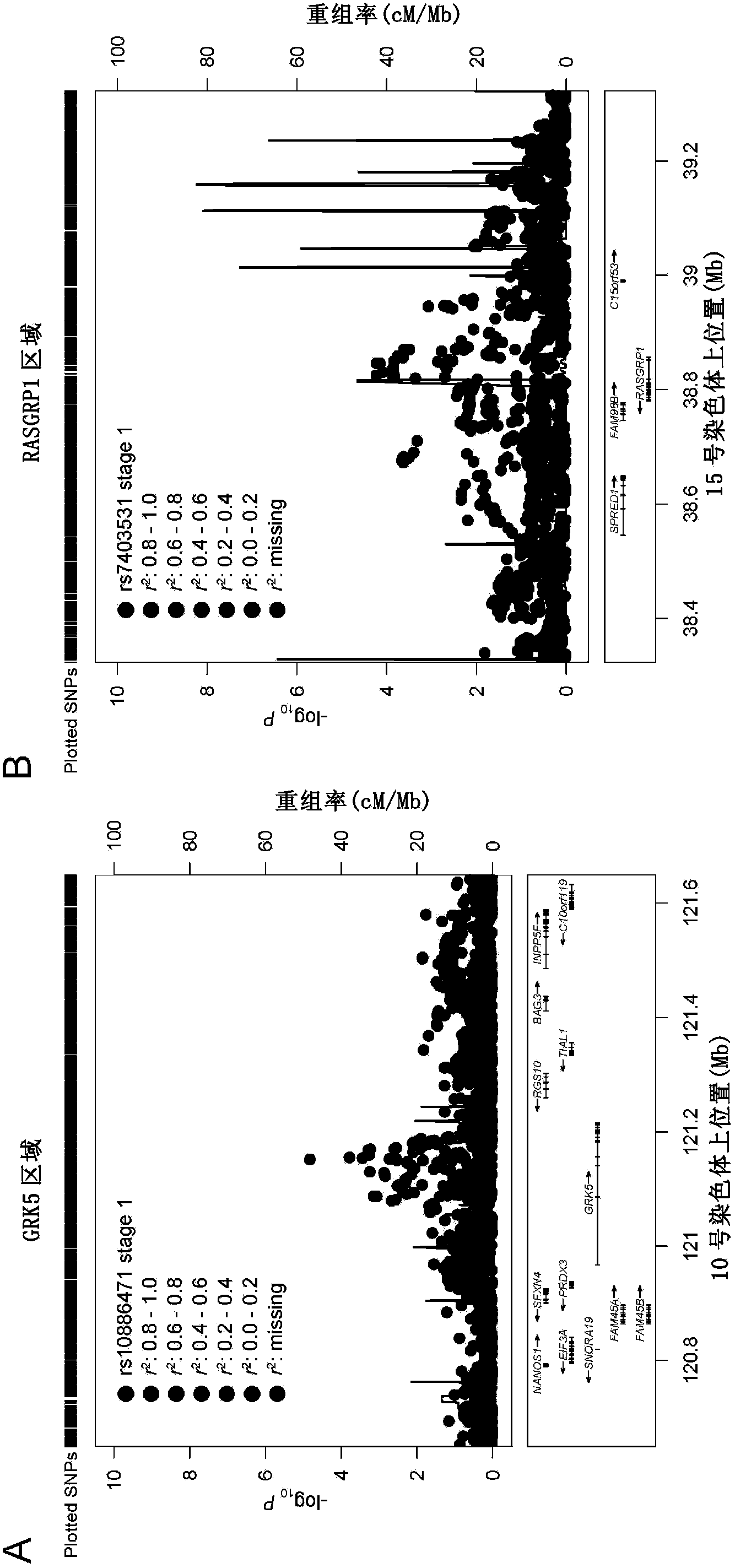
Type 2 diabetes susceptibility genetic locus as well as detection method and kit thereof
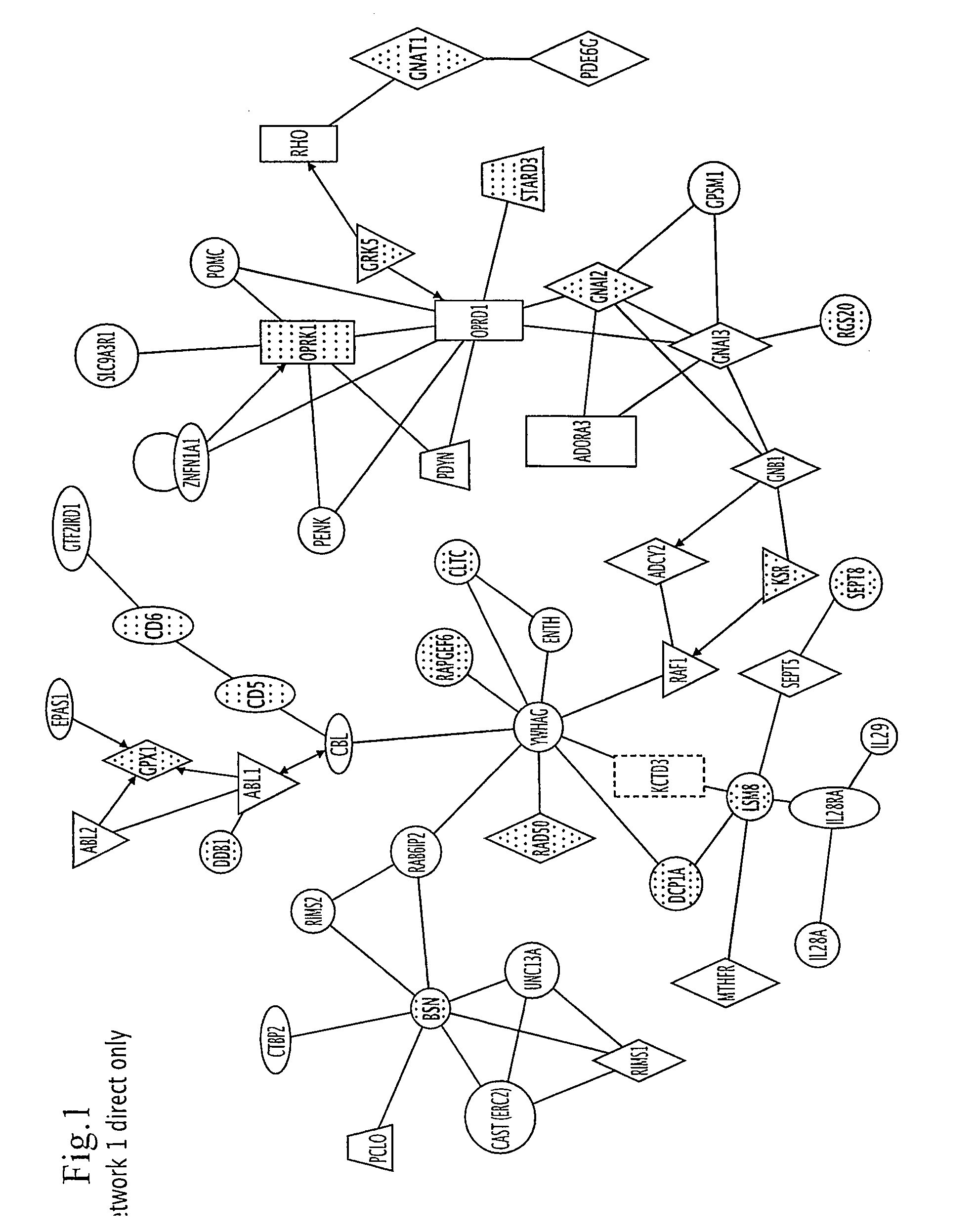
The invention relates to a type 2 diabetes susceptibility genetic locus as well as a detection method and a kit of the type 2 diabetes susceptibility genetic locus. Particularly, the invention discloses the type 2 diabetes susceptibility genetic locus found on the basis of genome-wide association study. The invention also provides the method for detecting the type 2 diabetes susceptibility. The method comprises steps of detecting individual GRK5 gene and RASGRP1 gene, and determining whether the transcript and / or protein has variation compared with normal transcript and / or protein, if yes, the individual is more likely to be attacked by the type 2 diabetes than normal people. The invention also discloses the corresponding kit for detection.
Owner:SHANGHAI INST OF BIOLOGICAL SCI CHINESE ACAD OF SCI
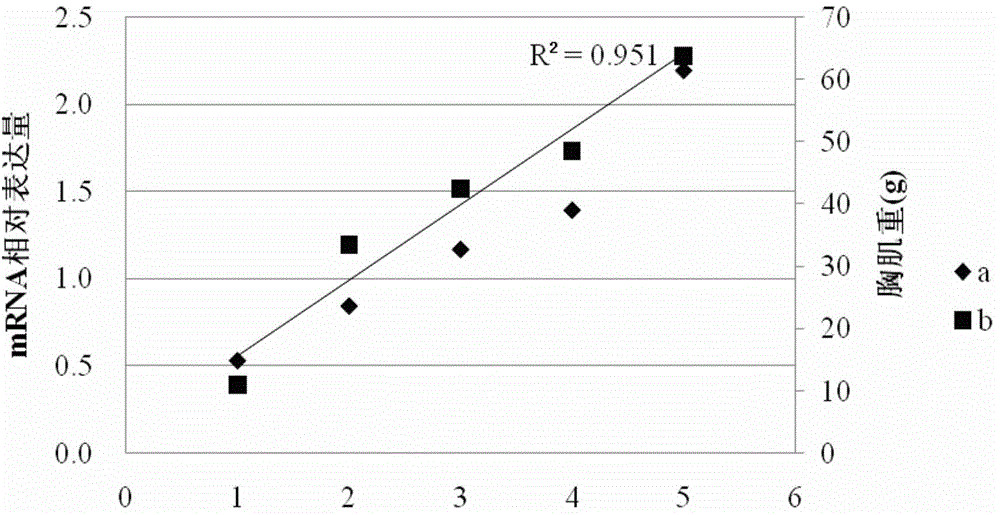
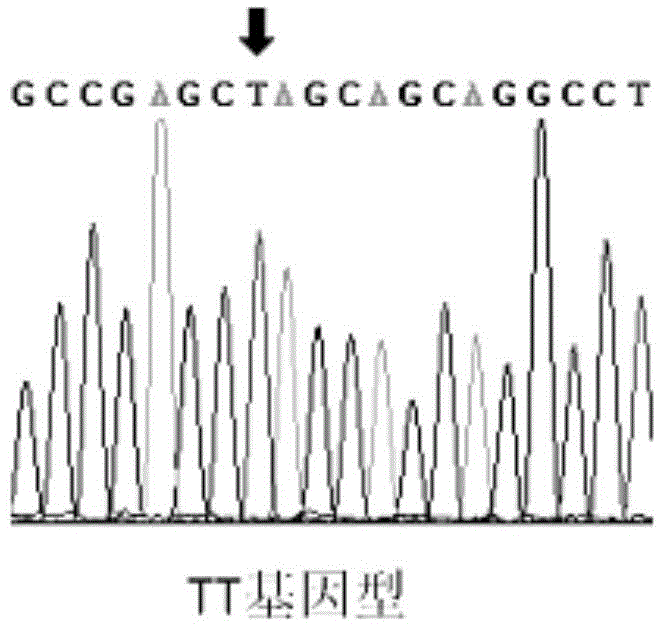
SNP molecular marker related to breast muscle weight and breast muscle percentage of chicken and application of SNP molecular marker
ActiveCN104673902AImprove breeding efficiencyImprove accuracyMicrobiological testing/measurementDNA/RNA fragmentationNucleotide sequencingGenetypes
The invention provides an SNP molecular marker related to the breast muscle weight and the breast muscle percentage of a chicken and application of the SNP molecular marker. The invention finds that the SNP molecular marker remarkably related to the breast muscle weight and the breast muscle percentage exists in a GJA1 gene on the basis of GWAS (Genome Wide Association Study). The invention provides a kit for detecting the SNP molecular marker; and the kit contains a pair of primers of which the nucleotide sequences are shown in SEQ ID NO. 1-2. The invention further provides a method for selecting a high-quality chicken pure line with high breast muscle weight and breast muscle percentage by utilizing the SNP site of the GJAI gene. The method comprises the following steps: obtaining a chicken genome DNA; detecting the genetype of the SNP T1289c site of the GJAI gene by virtue of the pair of primers of which the nucleotide sequences are shown in SEQ ID NO. 1-2; and selecting chicken individuals with superior genetype CC. The kit provided by the invention is simple to operate, high in flexibility, high in accuracy and low in detection cost and has an important application value.
Owner:INST OF ANIMAL SCI OF CHINESE ACAD OF AGRI SCI
SNP molecular marker related to daily gain of Duroc and application thereof
InactiveCN106636346AEasy to breedMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceGenome
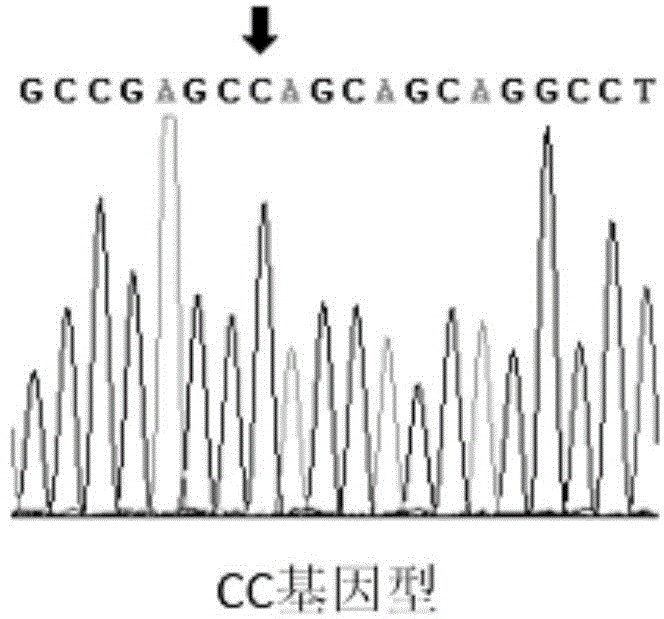
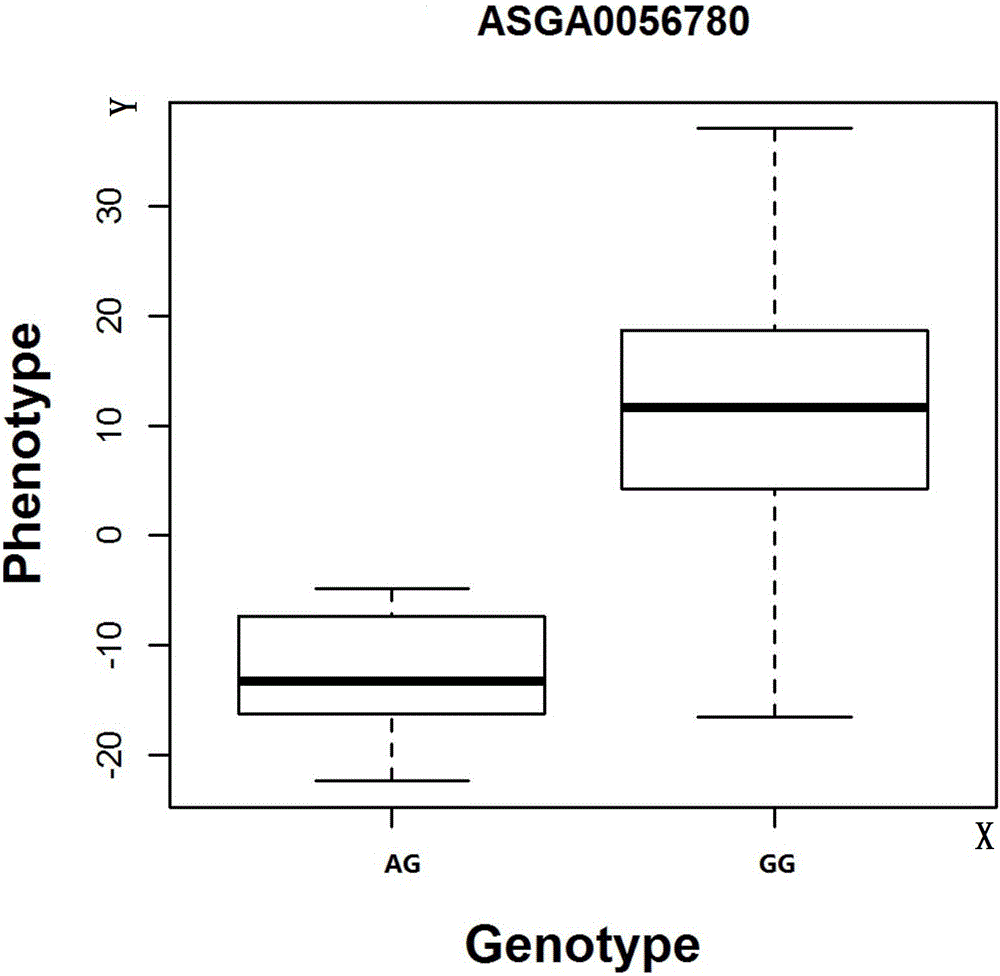
The invention discloses a SNP molecular marker related to daily gain of Duroc and an application thereof. Through genome-wide association study (GWAS), an SNP site (ASGA0056780) related to the daily gain of Duroc is identified at the No. 13 chromosome of a pig; the GG type individual at the site has faster daily gain. The invention also provides a method for detecting the SNP molecular marker at the same time; the Taq probe can judge the site rapidly. The method can exactly and rapidly classify the ASGA0056780 with low cost, and provide technical support to the establishment of the breeding method of the Duro type daily gain molecular marker.
Owner:广西柯新源原种猪有限责任公司 +6
Single nucleotide polymorphism (SNP) molecular markers related to pig effective teat pair number traits and application
ActiveCN104862388AMicrobiological testing/measurementDNA/RNA fragmentationNucleotideMolecular breeding
The invention belongs to the technical field of preparation of livestock molecular markers and particularly relates to single nucleotide polymorphism (SNP) molecular markers related to pig effective teat pair number traits and application. The SNP molecular markers are obtained through genome-wide association study, and nucleotide sequences of the molecular markers are shown in SEQ ID NO: 1-2 and figure 2-3 respectively. A corresponding haplotype that is shown in figure 4 is obtained through haplotype analysis by means of obtained SNP. The markers and the haplotype can be used for pig effective teat number molecular breeding.
Owner:HUAZHONG AGRI UNIV
Method for improving genome-wide prediction accuracy
ActiveCN110459265AImprove forecast accuracyImprove forecasting efficiencyProteomicsGenomicsNucleotideData information
The invention relates to the technical field of molecular breeding of crops and genome-wide association study, and particularly provides a method for improving the genome-wide prediction (GP) accuracy. The method comprises the steps that (1), a target crop population is subjected to phenotype and genotype identification, based on the genome-wide association study (GWAS) of the whole population, the four single nucleotide polymorphisms (SNPs) with the maximum effect are found; (2), the four SNPs with the maximum effect are used as the fixed effect, genotype and environment interaction components are added to a GP model, and the prediction accuracy can be improved to the greatest extent. After the phenotype and genotype identification is completed, the prediction accuracy is improved as faras possible only by using existing data information without increasing other investment of manpower and material resources. When the method is used in molecular breeding work, the prediction efficiency will be improved, therefore, the prediction reliability is improved, and the breeding cost is reduced.
Owner:INST OF CROP SCI CHINESE ACAD OF AGRI SCI
Molecular marker influencing feed conversion ratio character of pig and application
ActiveCN105624155ALower conversion rateReduce manufacturing costMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceFeed conversion ratio
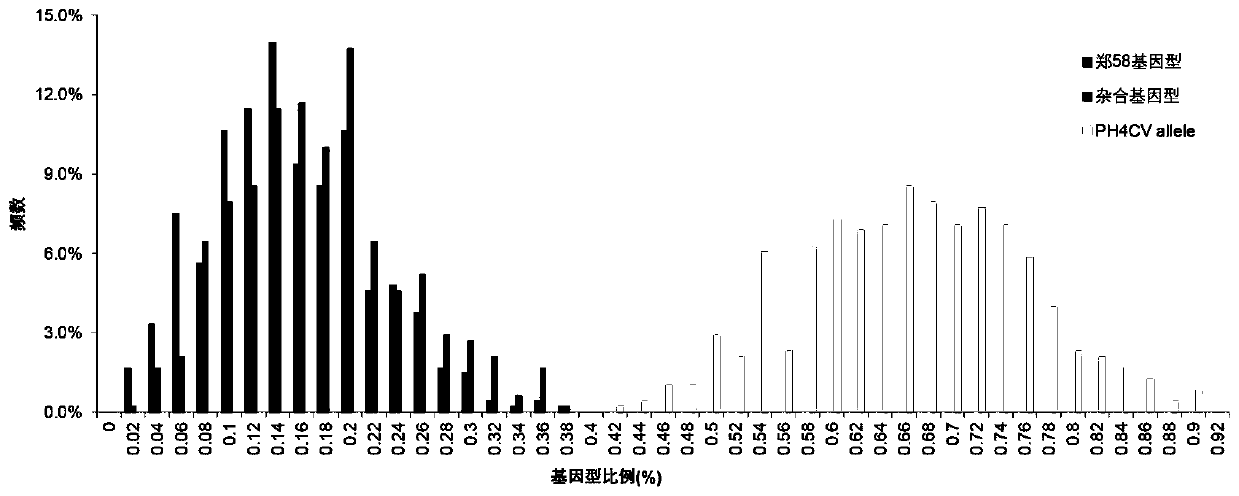
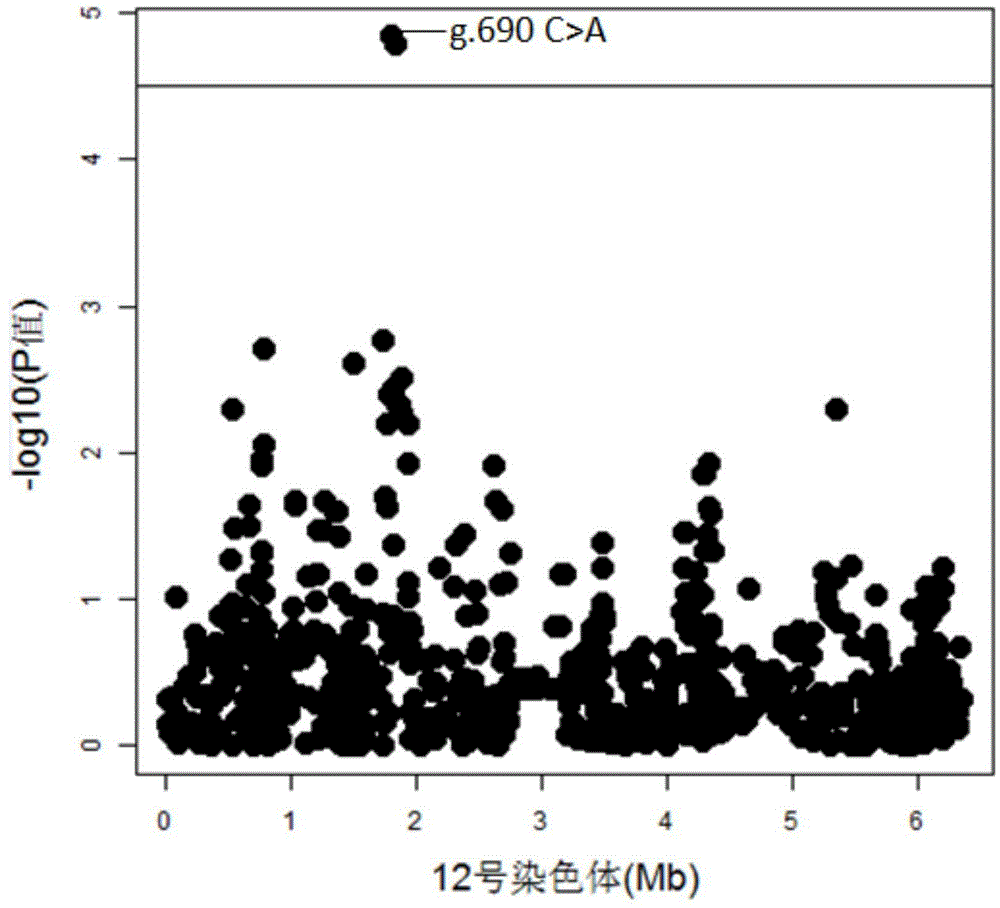
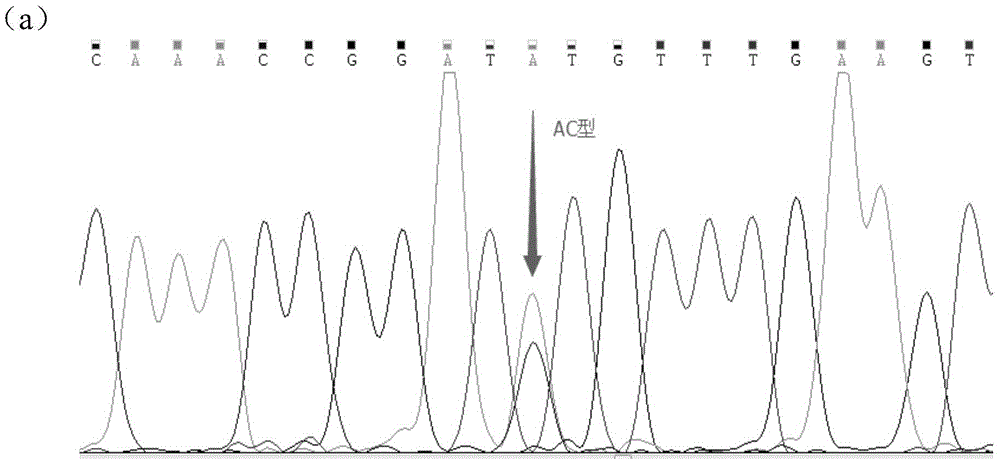
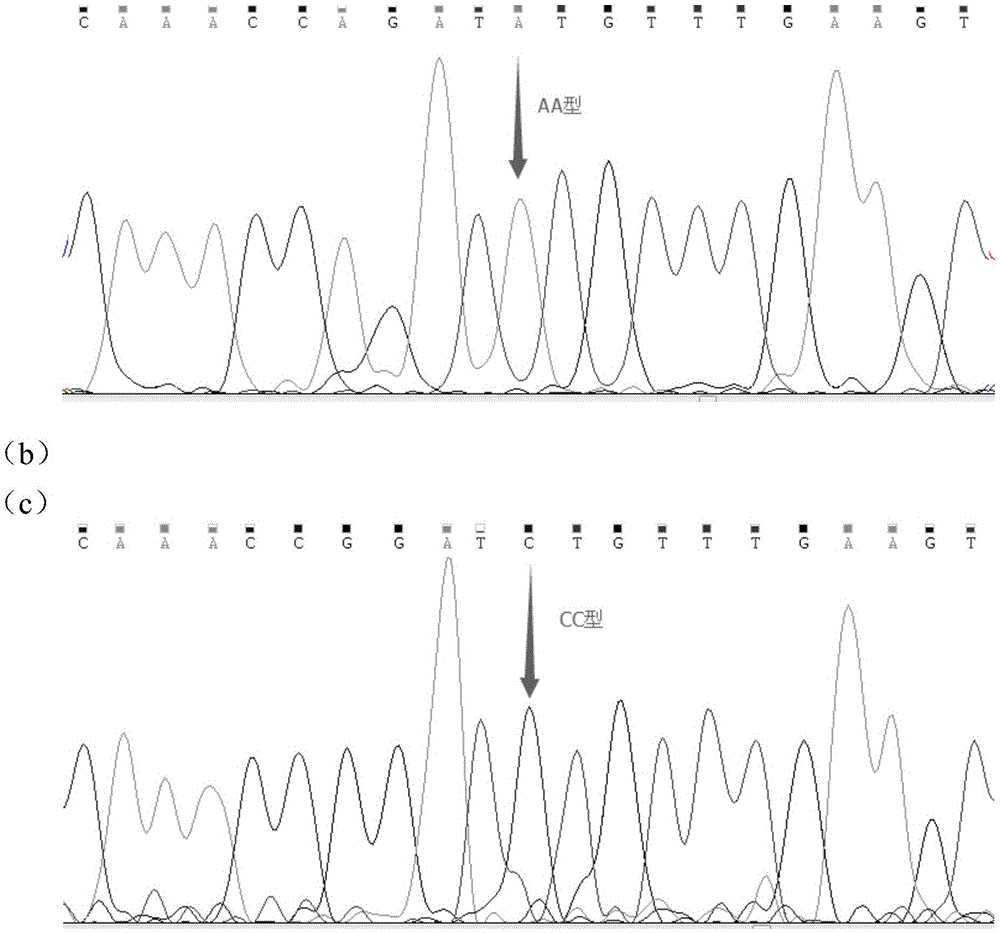
The invention discloses a molecular marker influencing the feed conversion ratio character of a pig and application, and belongs to the technical field of marker-assisted selection technologies and genetic breeding. The molecular marker is obtained through the genome-wide association study (GWAS), the nucleotide sequence of the molecular marker is shown in SEQ ID NO:1, the C>A nucleotide single-base mutation (named as g.690C>A) is located at the 690bpth position of the genomic sequence of the pig, and the feed conversion ratio character of the pig is remarkably influenced through mutation. The invention discloses preparation and application of the molecular marker.
Owner:SOUTH CHINA AGRI UNIV
Method for predicting complex diseases and phenotypic related metabolites based on Mendel randomization
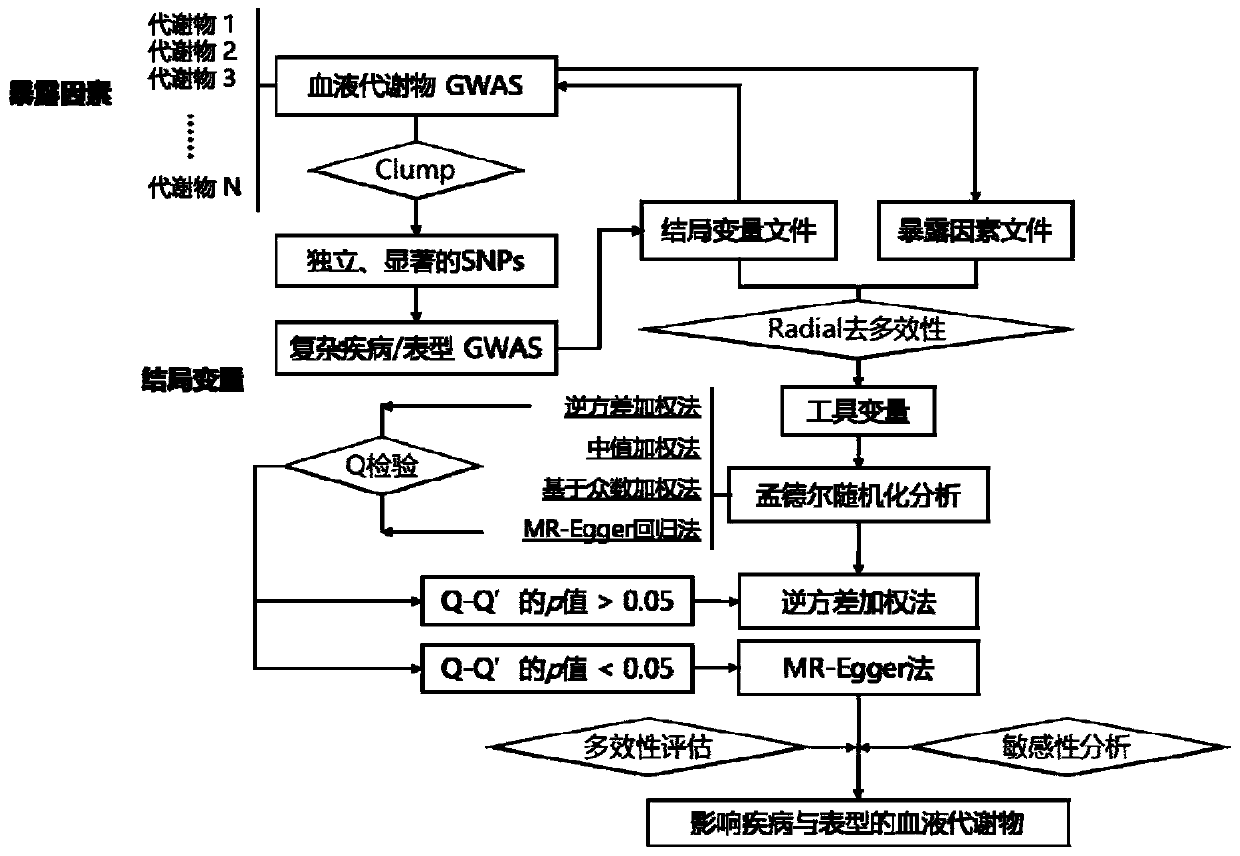
PendingCN111341448ARich in biological functionsImprove statistical powerHealth-index calculationProteomicsMetaboliteNutrition
The invention discloses a method for predicting complex diseases and phenotypic related metabolites based on Mendel randomization analysis. The method comprises the following steps: (1) collecting andsorting blood metabolite whole genome association research data; (2) screening tool variables and removing multiple effects; and (3) performing double-sample Mendel randomization analysis according to the screened tool variables, and obtaining metabolites related to diseases or phenotypes by integrating judgment of effect values and sensitivity analysis of multiple analysis methods. According tothe method, the influence of 363 blood metabolites on complex diseases and phenotypes is predicted through double-sample Mendel randomization analysis in combination with screening and discriminationof multiple methods, and an important foundation is laid for subsequent searching of drug targets for the blood metabolites and nutrition intervention.
Owner:XI AN JIAOTONG UNIV
Molecular marker for detecting anti-scab QTL (quantitative trait loci) Qfhb.hbaas-5AS and application method of molecular marker
ActiveCN109797241AMicrobiological testing/measurementDNA/RNA fragmentationAgricultural scienceQuantitative trait locus
The invention discloses a molecular marker for detecting anti-scab QTL (quantitative trait loci) Qfhb.hbaas-5AS and an application method of the molecular marker. The invention further provides application of a substance for detecting the genotype of SNP IWB21456 loci of wheat chromosomes 5AS in identification or auxiliary identification of the resistance of the wheat scab resistance, and application of the substance for detecting the genotype of the SNP IWB21456 loci of the wheat chromosomes 5AS in preparation of products for identification or auxiliary identification of the wheat scab resistance. Through genome-wide association study (GWAS), the loci Qfhb.hbaas-5AS on short arms of the wheat chromosomes 5AS are discovered, and are further correlated with the SNP IWB21456 to be convertedinto common PCR markers KASP-Fhb5AS, and the concordance rate of the two for 240 materials is 96.7%. The molecular marker can be used for detecting the genotype of the anti-scab QTL Qfhb.hbaas-5AS, and for anti-scab molecular breeding.
Owner:INST OF FOOD CROPS HUBEI ACAD OF AGRI SCI +1
Genetic diagnosis using multiple sequence variant analysis
The present invention is in the field of nucleic acid-based genetic analysis. More particularly, it discloses novel insights into the overall structure of genetic variation in all living species. The structure can be revealed with the use of any data set of genetic variants from a particular locus. The invention is useful to define the subset of variations that are most suited as genetic markers to search for correlations with certain phenotypic traits. Additionally, the insights are useful for the development of algorithms and computer programs that convert genotype data into the constituent haplotypes that are laborious and costly to derive in an experimental way. The invention is useful in areas such as (i) genome-wide association studies, (ii) clinical in vitro diagnosis, (iii) plant and animal breeding, (iv) the identification of micro-organisms.
Owner:METHEXIS GENOMICS
Application of Ghd7 gene in regulating rice flag leaf chlorophyll content
InactiveCN106148354AReduce chlorophyll contentFermentationGenetic engineeringAgricultural scienceChloroplast
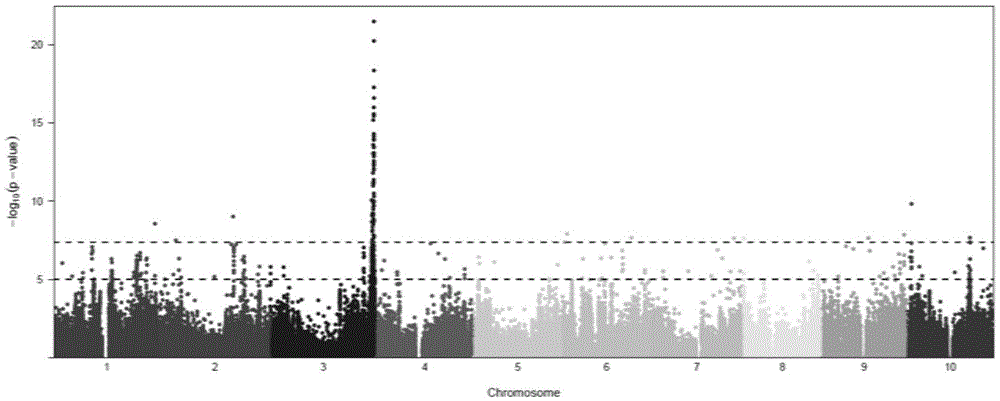
The invention belongs to the technical field of plant genetic engineering. It is found that a significant hot spot region exists in the range of 9.13 Mb to 9.17 Mb of a chromosome 7 through genome-wide association study (GWAS), nine signal peak values which have an extremely significant effect on the rice flag leaf chlorophyll content from different GWAS can be detected repeatedly, and the peak values correspond to Ghd7 gene positions. It is verified through near-isogenic lines and genetically modified materials that Ghd 7 serves as a main natural variation site to regulate the rice flag leaf chlorophyll content by down-regulating related gene expressions such as chlorophyll metabolism and chloroplast synthesis. The Ghd 7 has important breeding value in optimizing the rice flag leaf chlorophyll content.
Owner:HUAZHONG AGRI UNIV
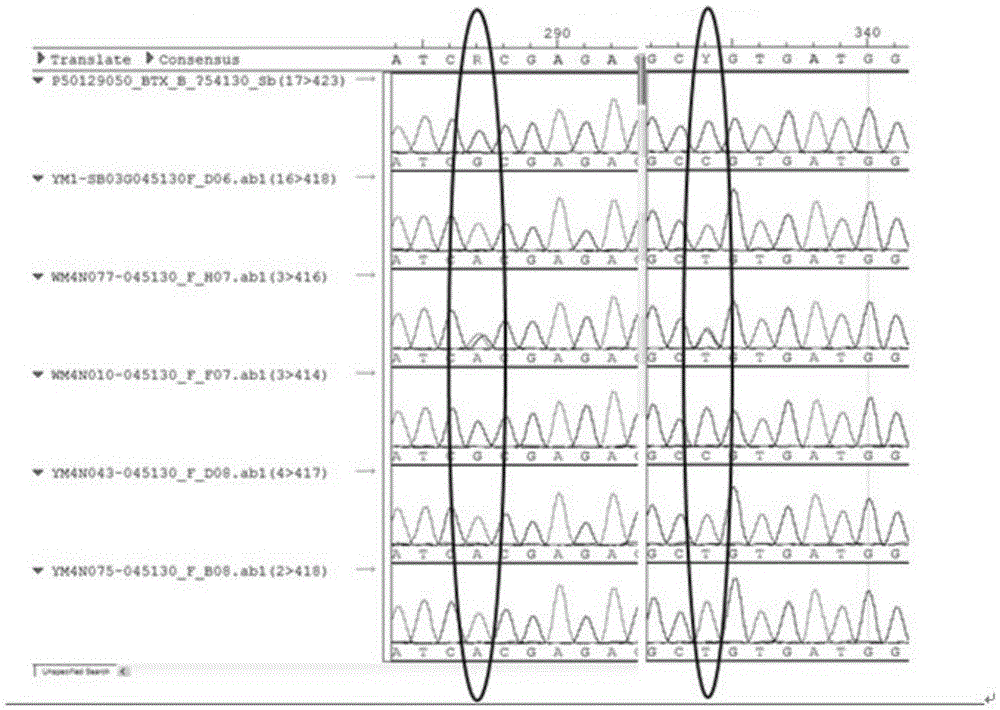
Gene SbAn-1 capable of controlling existence of sorghum awn and SNP mutation site and application
InactiveCN105603072AEasy to digAchieving Assisted BreedingMicrobiological testing/measurementDNA/RNA fragmentationPcr ctppSorghum
The invention discloses a gene SbAn-1 with the function of controlling existence of sorghum awn and belongs to the field of molecular biology and sorghum breeding. The gene SbAn-1 capable of controlling existence of sorghum awn is obtained through genome-wide association study. According to the technology, a plurality of nucleotide polymorphic sites can be detected at a time, and therefore large-batch excavation and simple trait genes are facilitated. The gene is selected on the basis of the PCR technology, and the technology can be directly used for identifying the gene SbAn-1 with the function of controlling existence of sorghum awn and allelic genes thereof, so that molecular marker assisted breeding is achieved.
Owner:LIAONING ACAD OF AGRI SCI +2
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com