Engineered CAS9 with wider DNA targeting range
A technology of N986R and R991L, applied in the field of engineered CAS9 protein, can solve problems such as limiting the targeting range and application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] This example demonstrates an all-atom molecular dynamics simulation of a SaCas9 complex.
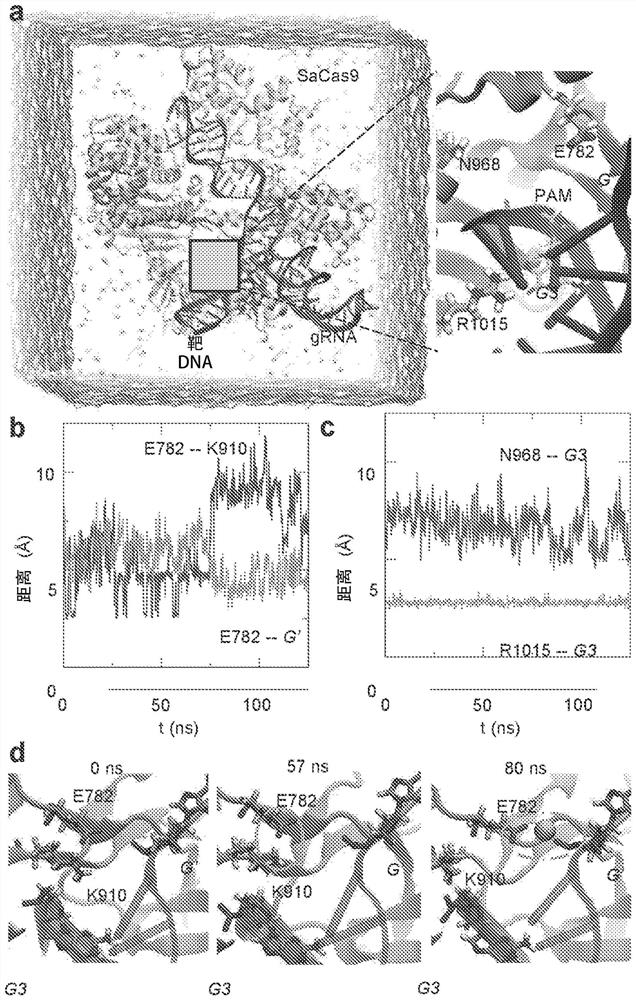
[0095] The high-resolution SaCas9 complex structure (Nishimasu et al., supra) was examined (although crystal contacts may affect the local structure; see figure 2 ). To establish the dynamic details of the SaCas9 complex in its native state, molecular dynamics (MD) methods were used to model the complex under physiological conditions. MD simulations as a complement to experimental studies have been shown to be effective in understanding protein-DNA interactions (Palermo et al., Proceedings of the National Academy of Sciences, 114:7260-7265 (2017); and Cong et al., Nat. Commun., 3:968 (2012)). All-atom MD simulations were performed to characterize the molecular mechanism of DNA target binding to the SaCas9-sgRNA complex (see figure 1 A).
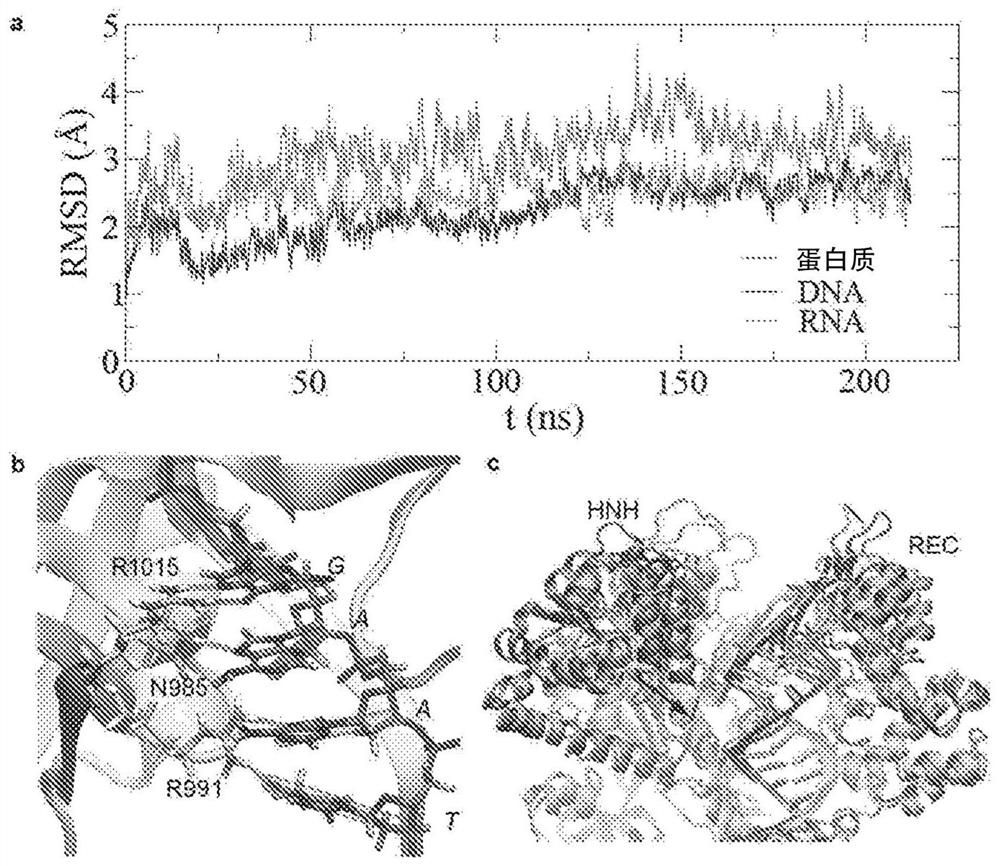
[0096] In MD analysis, after the binding state with the target DNA substrate reaches equilibrium ( image 3 A), The secondary structures ...
Embodiment 2
[0100] This example describes the use of free energy perturbations and experimental assays to probe Cas9 PAM recognition.
[0101] Completing the system equilibrium and MD analysis reveals previously underestimated dynamics of PAM recognition and addresses one of the fundamental challenges of modeling genome editing tools, namely how to understand in a quantitative manner the contribution of protein residues to target recognition. With this in mind, a combinatorial process was used, in which structural insights guided computational analysis followed by targeted gene editing experiments that further demonstrated computational mapping of Cas9 variant activity, where in silico predictions could correlate with experimental Cas9 editing efficiencies .
[0102] First, the contribution of PI domain residues to SaCas9 PAM recognition was quantified using free energy perturbation (FEP) calculations ( Figure 5 A). Alanine scanning analysis was performed directly on residues near the ...
Embodiment 3
[0104] This example describes the analysis of KKH SaCas9 variants to reveal the molecular mechanism of their expanded PAM.
[0105] The KKH mutant of SaCas9 involves three substitutions: E782K, N968K, and R1015H (Kleinstiver et al., Nature Biotech., 33:1293-1298 (2015). The thermodynamic cycle of R1015H is shown in Figure 5a. In the bound state, R1015 binds to G3 with two hydrogen bonds and is responsible for the PAM-specific NNGRRT. This interaction is further stabilized by the salt bridge between E993 and R1015, which can significantly reduce the conformational fluctuation of R1015. Such as Figure 5 As shown in A, the same salt bridge also exists in the free state of SaCas9. After the R1015H mutation, H1015 in the bound state moves away from G3, thereby releasing the specificity for G3 in the NNGRRT PAM. However, this mutation (in Figure 5 Represented by ΔG1 in A) significantly reduces the binding free energy (or binding affinity). The same mutation process as in th...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap