A method and system for automatic pairing of gene sequencing multi-sample data files
A data file and gene sequencing technology, applied in the field of information processing, can solve the problems of huge size, difficult management of data file file names, and easy modification of file names by humans, so as to facilitate management, improve use efficiency, and reduce program execution errors. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1. Paired gene sequencing multi-sample data files by the method of the present invention
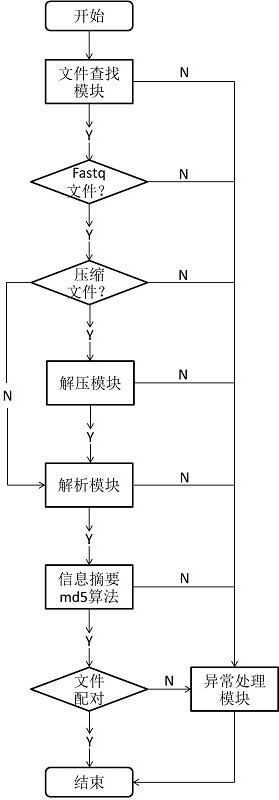
[0041] 1) The file search module interprets whether it is a file, then traverses all files, parses the file name, and the suffix characters of the file name include fq and fastq, which are the FASTQ files that need to be processed, and other files will not be processed.
[0042] 2) The file decompression module judges that the files whose file name suffixes are .gz, .zip, and .bz are compressed files, and need to be decompressed and decompressed into text files, and other files are not processed. During the processing, if an exception occurs, it will be handled by the exception handling module.
[0043] 3) The file reading module reads the content of the FASTQ file line by line, and removes special characters such as spaces and carriage returns at the beginning and end of the line. During the processing, if an exception occurs, it will be handled by the exception handling...
experiment example 1
[0050] Experimental example 1, the automatic pairing effect of the method of the present invention
[0051] In the / fastq1 directory, there are 2900 FASTQ data files of 1450 samples. The traditional method is to manually organize the paired data files of each sample by file name, which takes at least 1 hour.
[0052] Using the method system for automatic pairing in Example 1 of the present invention, the file can be quickly matched and output, and the identification of the template chain and the complementary chain can be made. The filename prefixes of the two paired data files are the same, and the suffix contains R1 characters. file, the suffix containing the R2 character is the complementary chain data file. The whole process only takes less than 3 seconds, and at the same time, two errors are effectively found, namely, the data file cannot be matched and the file decompression error.
[0053] It can be seen that the method of the present invention can accurately, effectiv...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com