Use of self-assembled monolayers to probe the structure of a target molecule
a self-assembled monolayer and target molecule technology, applied in the field of self-assembled monolayers to probe the structure of a target molecule, can solve the problems of low affinity interaction that cannot adequately compete with larger, more diverse natural ligands, complex and expensive synthesis schemes, etc., and achieve the effect of increasing the binding affinity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
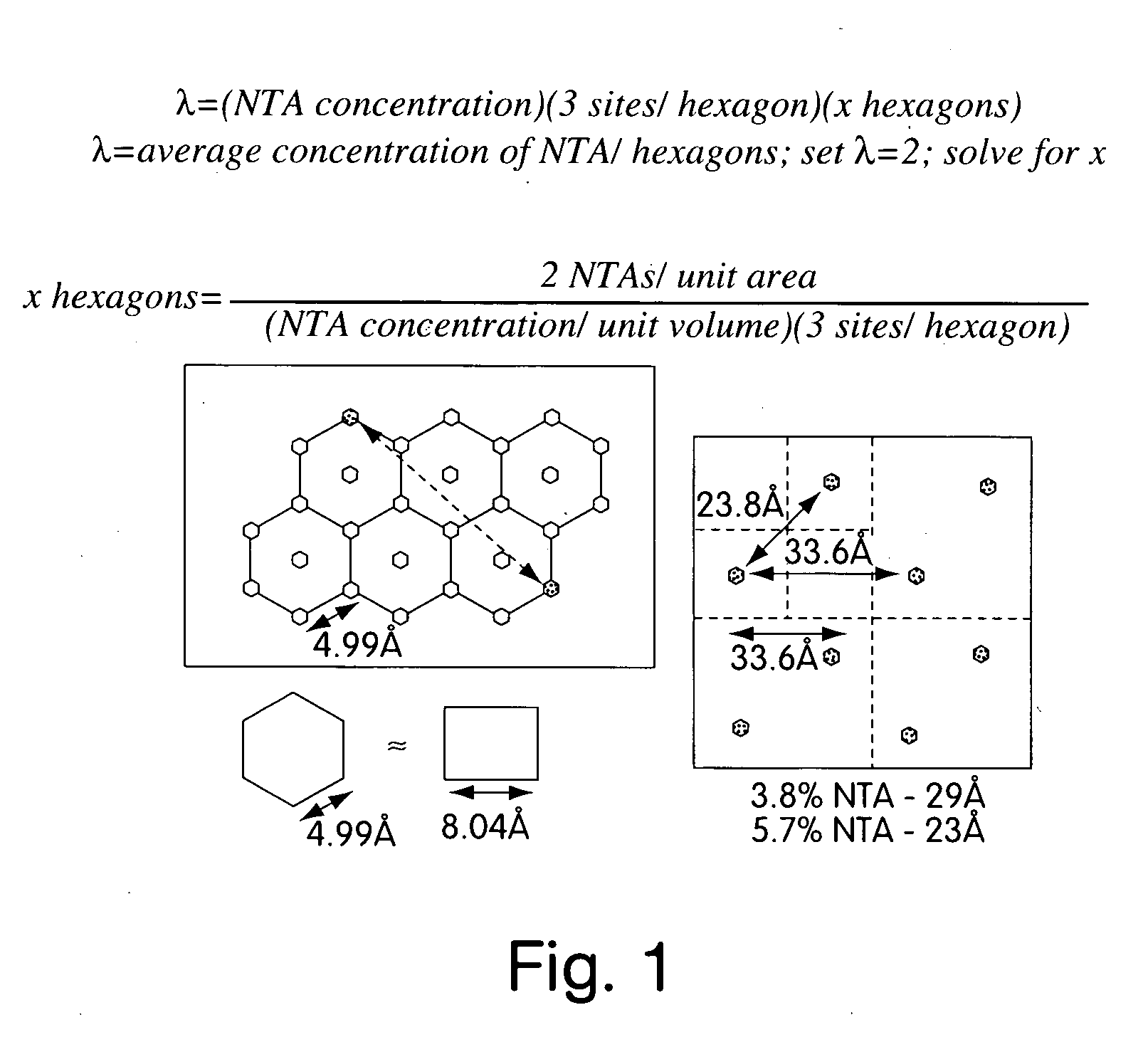
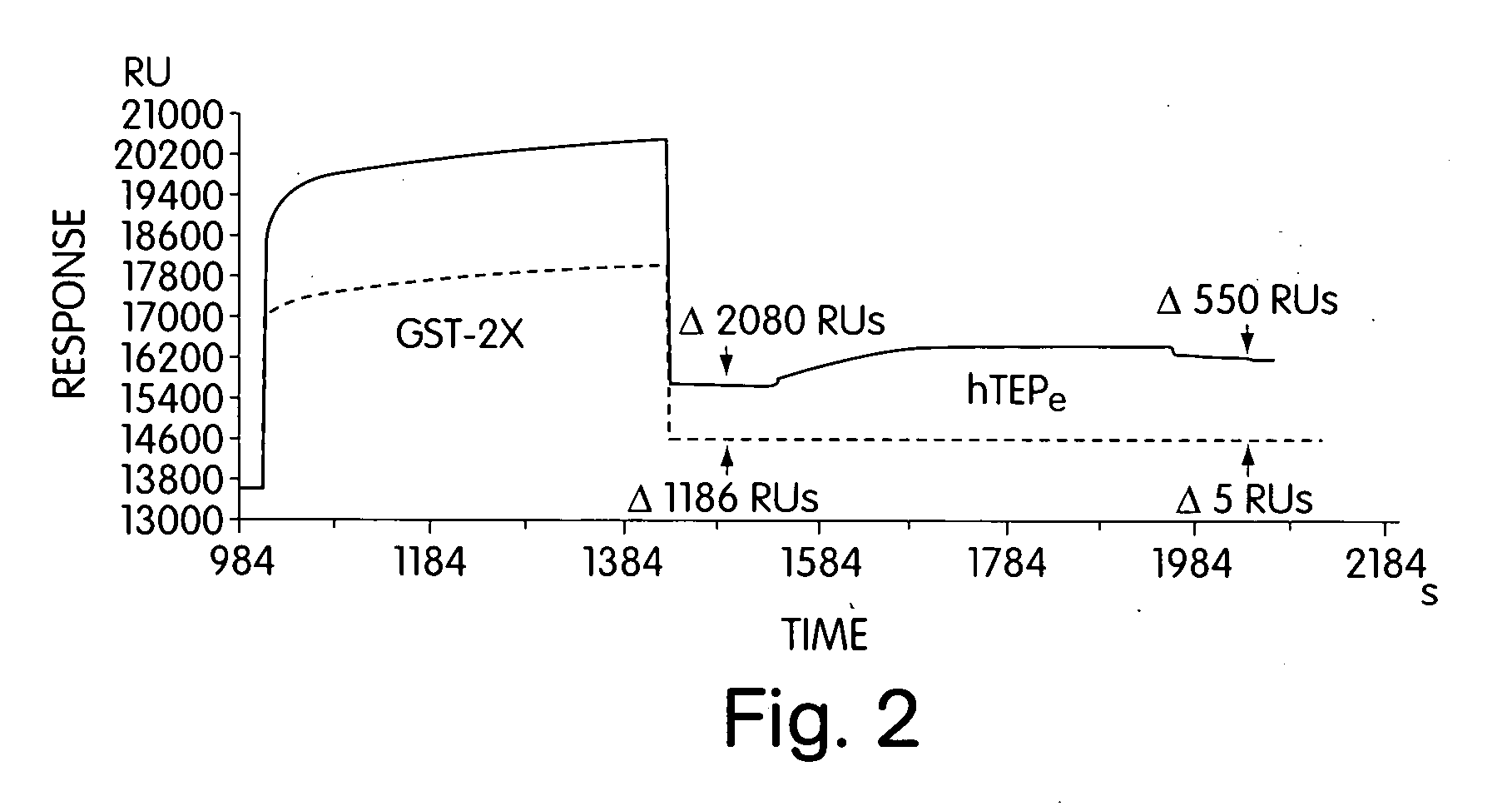
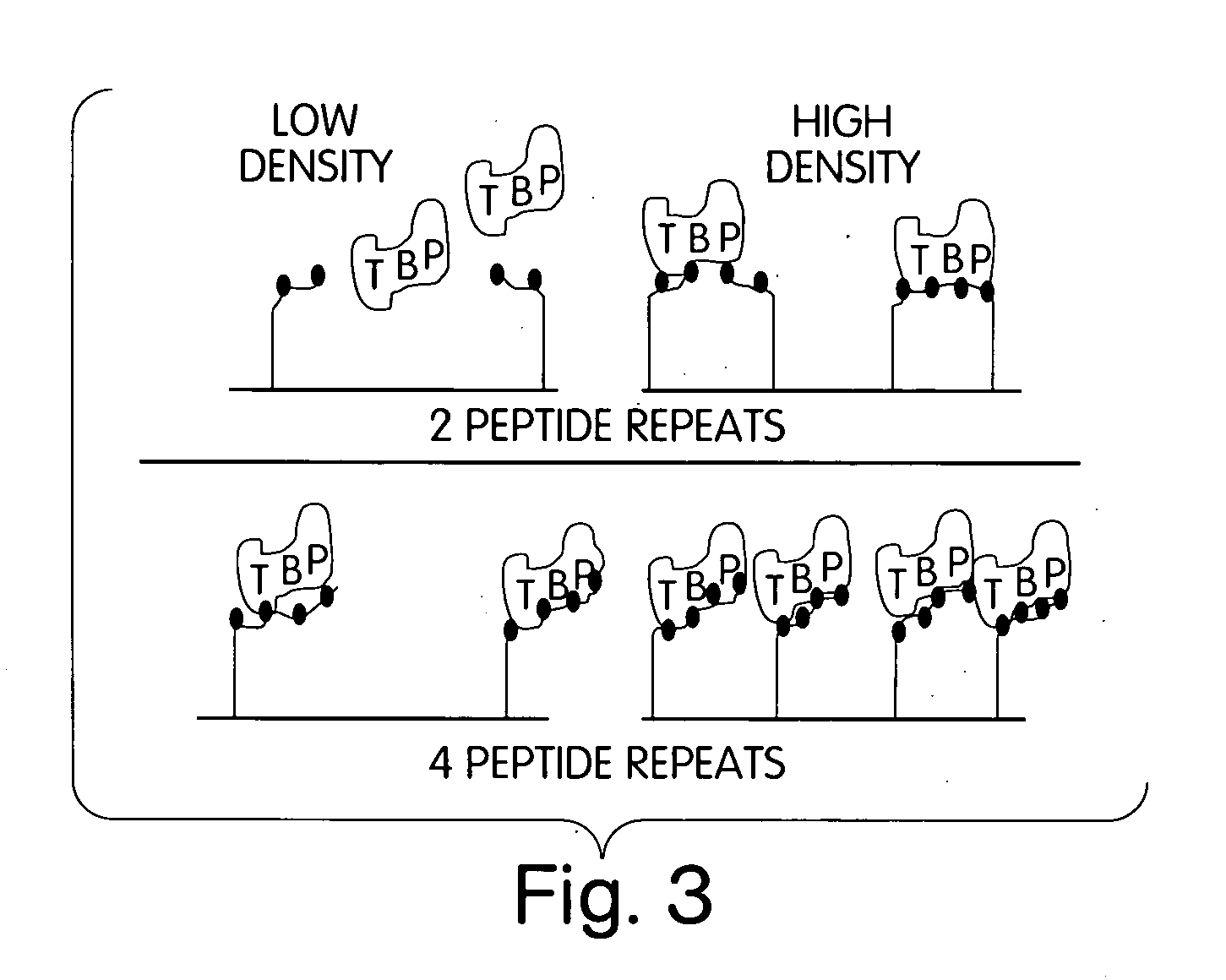
[0019] Variable density nitrilotriacetic acid (NTA)-SAMs were used to probe the binding site(s) of a biologically important molecule, the human general transcription factor TATA box binding protein (hTBP) [Burley, S. K. and Roeder, R. G. (1996) Biochemistry and structural biology of transcription factor IID (TFIID). Annu. Rev. Biochem. 65:769-799]. This transcription factor has been implicated as a direct target of transcriptional activators such as VP16 [Ingles, J. C., M. Shales, W. D. Cress, S. J. Triezenberg and J. Greenblatt. (1991) Reduced binding of TFIID to transcriptionally compromised mutants of VP16. Nature. 351:588-590]. In fact, the need for an activator is eliminated when TBP is artificially tethered to a DNA promoter [Xiao, H., J. D. Friesen and J. T. Lis. 1995. Recruiting TATA-binding protein to a promoter: transcriptional activation without an upstream activator. Mol. and Cell. Biol. 15(10):5757-5761].
[0020] Transcriptional activator proteins are modular in that the...
PUM
| Property | Measurement | Unit |
|---|---|---|
| flow rate | aaaaa | aaaaa |
| affinity | aaaaa | aaaaa |
| structure | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com