Fatty acid elongases
a technology of elongase and fatty acids, which is applied in the field of fatty acid elongase complexes and nucleic acids encoding elongase proteins, can solve the problems of hampered elucidation of the biochemistry of elongase complexes, hampered elucidation of the biochemistry of kas enzymes, and unsuitable for edible oils, etc., to achieve the effect of alter
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Cloning and Expression of FAE1 in Yeast Cells
[0061] The open reading frame of the Arabidopsis FAE1 gene was amplified directly by PCR, using Arabidopsis thaliana cv. Columbia genomic DNA as a template, pfu DNA polymerase and the following primers: 5′CTCGAGGAGCAATGACGTCCGTTAA-3′ and 5′-CTCGAGTTAGGACCGACCGTTTTG-3′. The PCR product was blunt-end cloned into the Eco RV site of pBluescript (Stratagene, La Jolla, Calif.),
[0062] The FAE1 gene was excised from the Bluescript vector with BamH1, and then subcloned into the pYEUra3 (Clontech, Palo Alto, Calif.). pYEUra3 is a yeast centromere-containing, episomal plasmid that is propagated stably through cell division. The FAE1 gene was inserted downstream of a GAL1 promoter in pYEUra3. The GALL promoter is induced when galactose is present in the medium and repressed when glucose is present in the growth medium.
[0063] Insertion of the FAE1 gene in the sense orientation was confirmed by PCR, and pYEUra3 / FAE1 was used to transform Saccharomyc...
example 2
FAE1 Activity in Yeast Microsomes
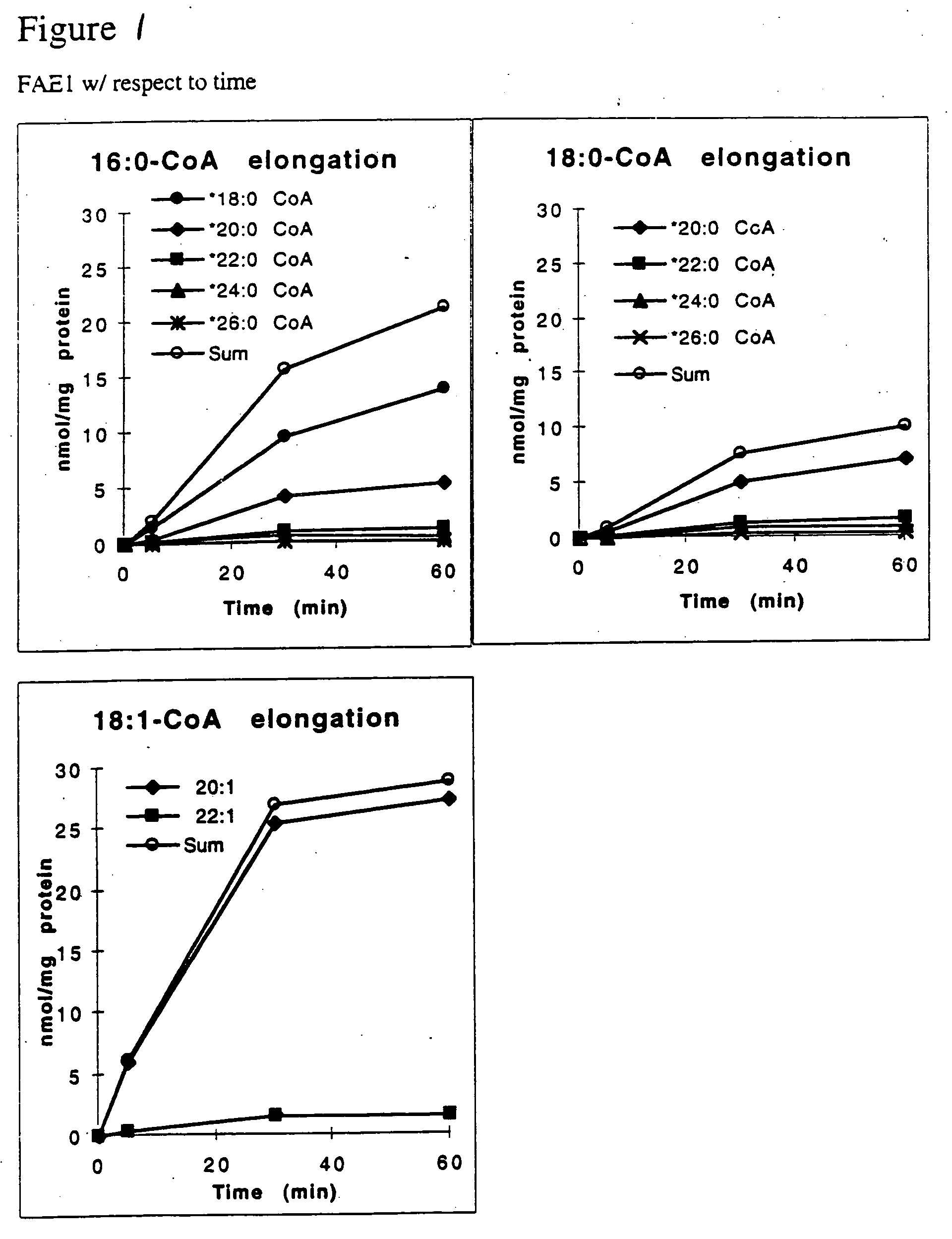
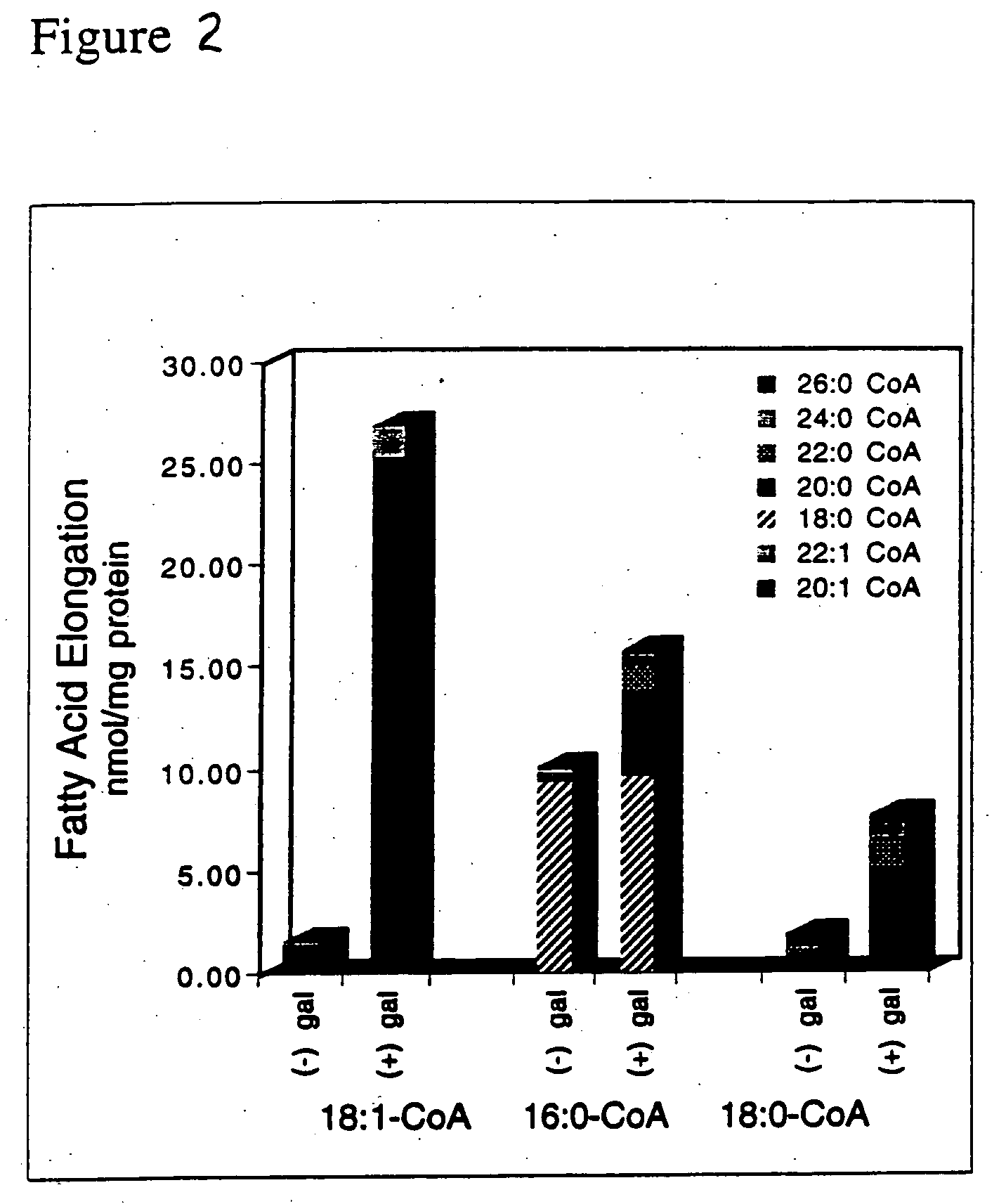
[0068] The functional expression of the FAE1 KAS was analyzed by isolating microsomes from transformed yeast cells and assaying these microsomes in vitro for elongase activity.
[0069] Transformed yeast cells were grown in the presence of either glucose or galactose (2% w / v) as described in Example 1. Cells were harvested by centrifugation at 5000×g for 10 min and washed with 10 ml ice cold isolation buffer (IB), which contains 80 mM Hepes-KOH, pH 7.2, 5 mM EGTA, 5 mM EDTA, 10 mM KCl, 320 mM sucrose and 2 mM DTT). Cells were then resuspended in enough IB to fill a 1.7 ml tube containing 700 μl of 0.5 μm glass beads and yeast microsomes were isolated from the cells essentially as described in Tillman, T. and Bell, R., J. Biol. Chem. 261:9144-9149 (1986). The microsomal membrane pellet was recovered by centrifugation at 252,000×g for 60 min. The pellet was rinsed by resuspending in 40 ml fresh IB and again recovered by centrifugation at 252000×g for 60...
example 3
Cloning and Sequencing of Arabidopsis Elongase Genes
[0076] The sequence of a jojoba seed cDNA (see WO 93 / 10241 and WO 95 / 15387, incorporated herein by reference) was used to search the Arabidopsis expressed sequence tag (EST) database of the Arabidopsis Genome Stock Center (The Ohio State University, Columbus, Ohio). The BLAST computer program (National Institutes of Health, Bethesda, Md., USA) was used to perform the search. The search identified two ESTs (ATTS1282 and ATTS3218) that had a high degree of sequence identity with the jojoba sequence. The ATTS1282 and ATTS3218 ESTs appeared to be partial cDNA clones rather than full-length clones based on the length of the jojoba sequence.
[0077] A genomic DNA library from Arabidopsis thaliana cv. Columbia, was prepared in the lambda GEM11 vector (Promega, Madison, Wis.) and was obtained from Ron Davis, Stanford University, Stanford, Calif. The library was hybridized with ATTS1282 and ATTS3218 as probes and 2 clones were identified fo...
PUM
| Property | Measurement | Unit |
|---|---|---|
| ionic strength | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com