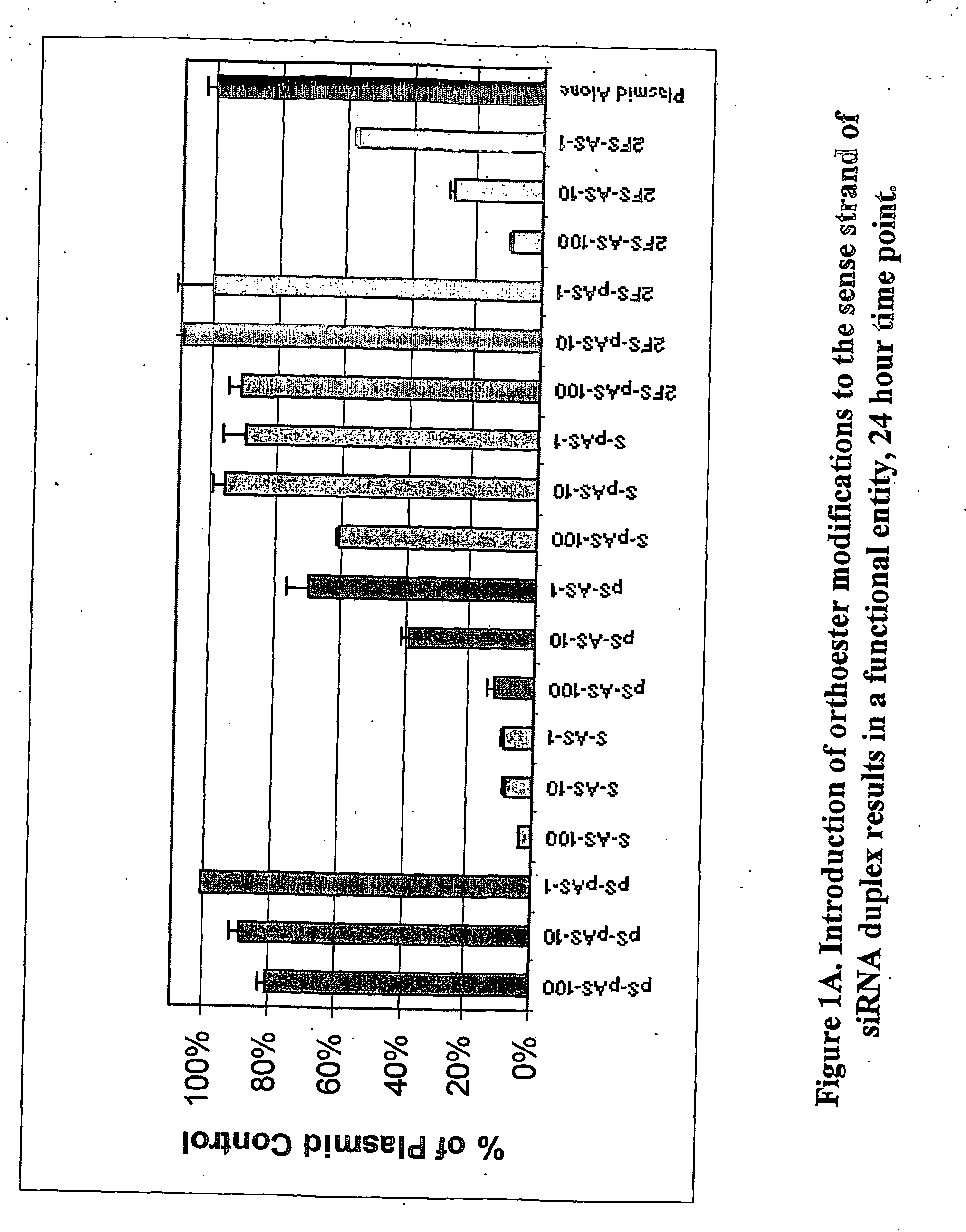
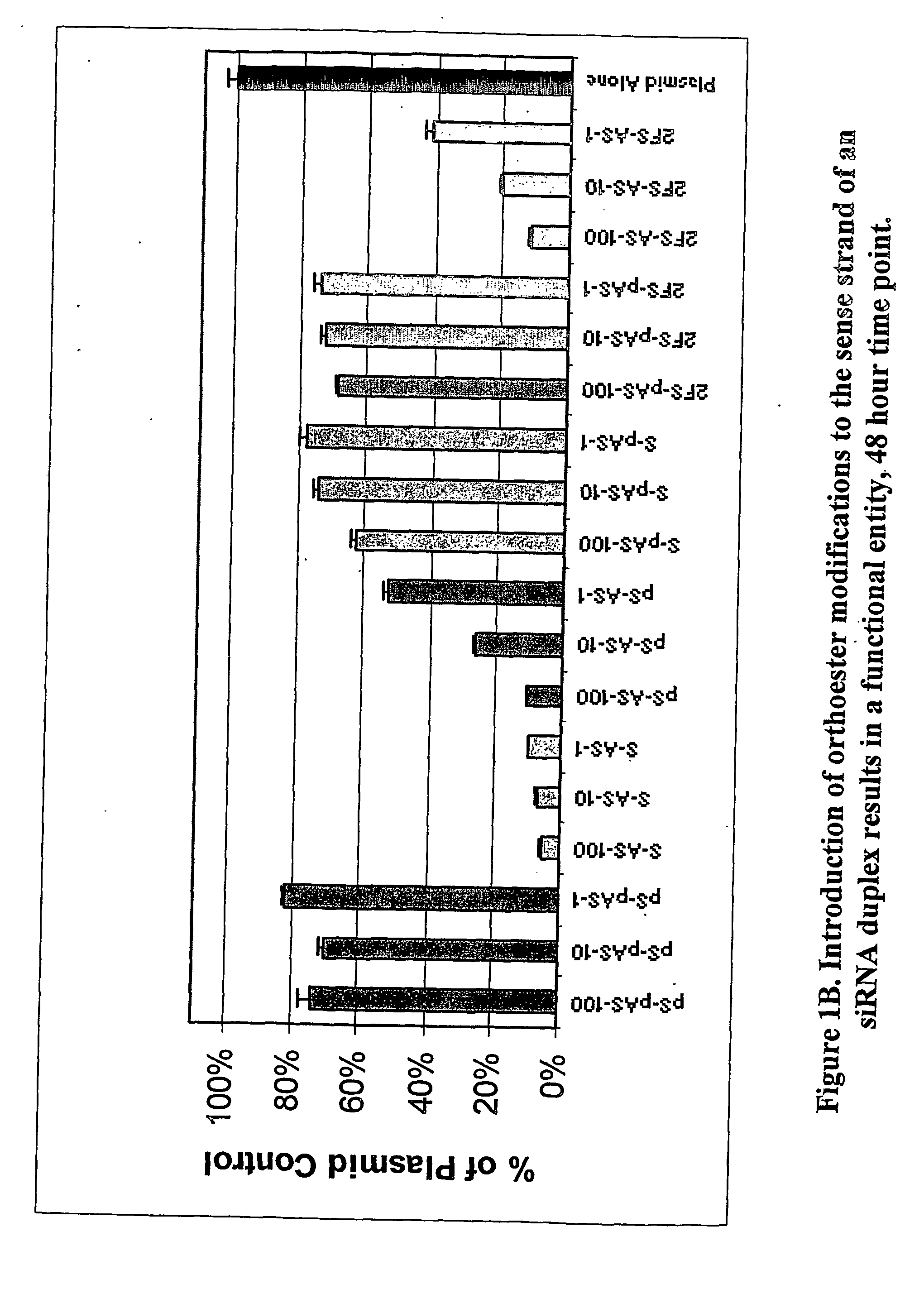
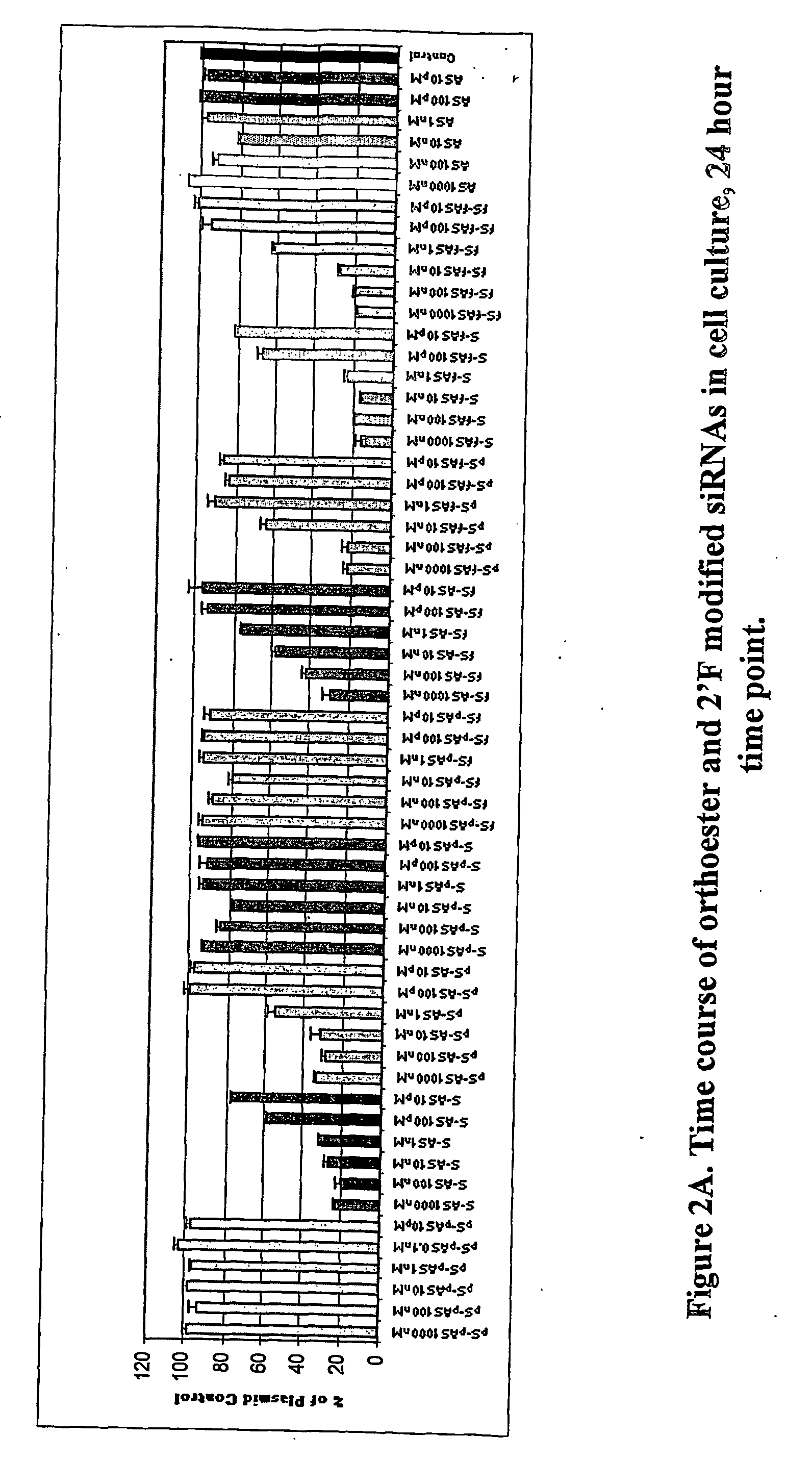
Modified polynucleotides for use in rna interference
a technology of rna interference and polynucleotides, which is applied in the direction of biocide, transferases, genetic material ingredients, etc., can solve the problems of reducing the stability of rnai, reducing the efficiency of rnai, so as to improve the stability, enhance stability, and maintain biological functionality.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesizing siRNAs
[0418] RNA oligonucleotides were synthesized in a stepwise fashion using the nucleotide addition reaction cycle illustrated in FIG. 13. The synthesis is preferably carried out as an automated process on an appropriate machine. Several such synthesizing machines are known to those of skill in the art. Each nucleotide is added sequentially (3 ′- to 5′-direction) to a solid support-bound oligonucleotide. Although polystyrene supports are preferred, any suitable support can be used. The first nucleoside at the 3 ′-end of the chain is covalently attached to a solid support. The nucleotide precursor, an activated ribonucleotide such as a phosphoramidite or H-phosphonate, and an activator such as a tetrazole, for example, S-ethyl-tetrazote (although any other suitable activator can be used) are added (step i in FIG. 13), coupling the second base onto the 5′-end of the first nucleoside. The support is washed and any unreacted 5′-hydroxyl groups are capped with an acetyla...
example 2
Deprotection and Cleavage of Synthesized Oligos From the Support
[0432] Cleaving can be done manually or in an automated process on a machine. Cleaving of the protecting moiety from the internucleotide linkage, for example a methyl group, can be achieved by using any suitable cleaving agent known in the art, for example, dithiolate or thiophenol. One molar dithiolate in DMF is added to the solid support at room temperature for 10 to 20 minutes. The support is then thoroughly washed with, for example, DMF, then water, then acetonitrile. Alternatively a water wash followed by a thorough acetonitrile will suffice to remove any residual dithioate.
[0433] Cleavage of the polynucleotide from the support and removal of exocyclic base protection can be done with 40% aqueous N-methylamine (NMA), followed by heating to 55 degrees Centigrade for twenty minutes. Once the polynucleotide is in solution, the NMA is carefully removed from the solid support. The solution containing the polynucleotid...
example 3
siRNAs Synthesized for Use in RNA Interference
[0438] The following is a list of 19-mer siRNAs having a di-dT overhang that were synthesized using Dharmacon, Inc.'s proprietary ACE chemistry, and were designed and used in accordance with the invention described herein. “SEAP” refers to human secreted alkaline phosphatase; “human cyclo” refers to human cyclophilin; an asterisk between nucleotide units refers to a modified internucleotide linkage that is a phosphorothioate linkage; the structure 2′-F—C or 2′-F-U refers to a nucleotide unit having a fluorine atom attached to the 2′ carbon of a ribosyl moiety; the structure 2′-N—C or 2′-N-U refers to a nucleotide unit having an —NH2 group attached to the 2′ carbon of a ribosyl moiety; the structure 2′-OME-C or 2′-OME-U refers to a nucleotide unit having a 2′-O-methyl modification at the 2′ carbon of a ribosyl moiety of either Cs or Us, respectively; dG, dU, dA, dC, and dT refer to a nucleotide unit that is deoxy with respect to the 2′ p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| half life | aaaaa | aaaaa |
| half life | aaaaa | aaaaa |
| v/v | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com