Detection of Fusion Events Using Replicate PCR Reactions
a technology of fusion events and pcr, applied in the field of gene fusion events, can solve the problems of only being able to detect gene fusion, fish cannot be applied to cells or tissues, and the approach cannot be applied to mrna
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
of EML4-ALK Variant at a Range of Allelic Fractions
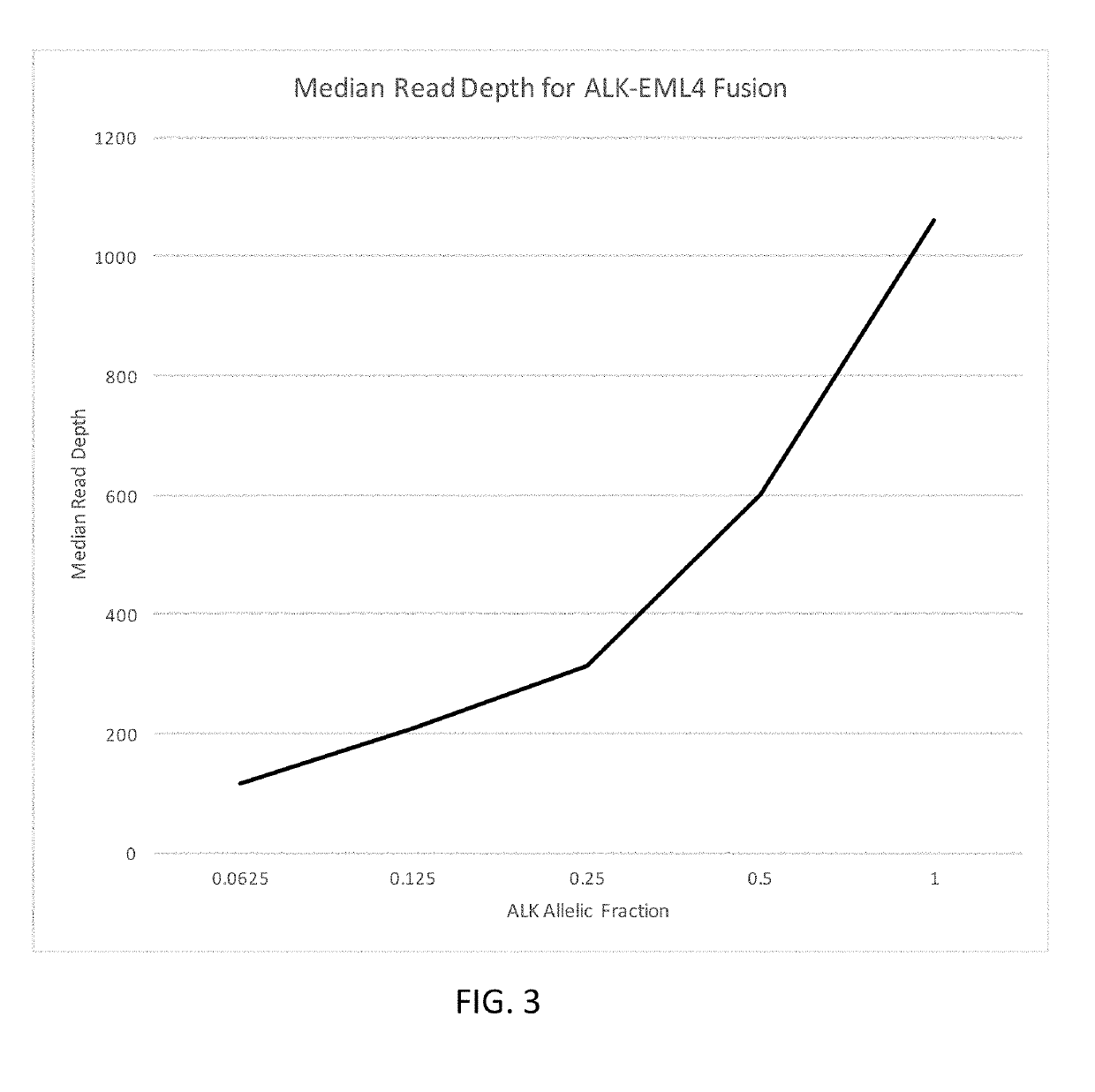
[0499]A custom cell free DNA reference standard containing an EML4-ALK fusion of sequence GAAGTTCCTATACTTTCTAGAGAATAGGAACTTC (SEQ ID NO: 1) at an allelic fraction of 2.5% was obtained from Horizon Discoveries. This reference standard was diluted in sheared (average 188 bp) human placental DNA (Bioline) to achieve allelic fractions of 1%, 0.5%, 0.25%, 0.125% and 0.0625%. Three samples were created at each allelic fraction.
[0500]Each sample was split into two replicates, each containing a total of 4000 input copies. PCR amplification was performed on two replicates using the ALK primer panel (table 1). Each PCR contained 25 uL DNA, 27.5 uL Platinum SuperFi 2× Master Mix (Invitrogen) and 2.5 uL of the ALK primer pool (for primer concentration see table 1). PCR cycling was followed using manufacturer' instructions. The PCR product was cleaned up using SPRIselect reagent (Beckman Coulter B23319) using the manufacturers protocol. DNA was ...
example 2
of ROS1-CD74 Variant
[0502]A synthetic gBlock containing a ROS1 fusion sequence (based on a sequence reported in the literature: Seki, Mizukami and Kohno, Biomolecules, 2015, 5, 2464-2476) was synthesized by IDT and was sheared using the covaris to achieve an average size of 150 bp. The gBlock was added to sheared (average 188 bp) human placental DNA (Bioline) to achieve an allelic fraction of 1%.
[0503]Each sample was split into two replicates, each containing a total of 4000 input copies. PCR amplification was performed on two of the replicates using the ROS1 primer panel (table 2). Each PCR contained 25 uL DNA, 27.5 uL Platinum SuperFi 2× Master Mix (Invitrogen) and 2.5 uL of the ROS1 primer pool (for primer concentration see table 2). PCR Cycling was followed using manufactures instructions. The PCR product was cleaned up using SPRIselect reagent (Beckman Coulter B23319) using the manufacturers protocol. DNA was eluted in 18 uL and a second PCR using Indexed illumina primers was p...
example 3
of ROS1-CD74 Variant Using Sequential Amplification
[0505]The same synthetic ROS1 fusion gBlock at 1% allelic fraction as was used in Example 2 was tested. Each sample was split into two replicates, each containing a total of 4000 input copies. Linear amplification of the template was performed on two of the replicates using only the ROS1 forward primer panel. Each reaction contained 25 uL DNA, 27.5 uL Platinum SuperFi 2× Master Mix (Invitrogen) and 2.5 uL of the ROS1 forward primer pool. Cycling was followed using manufactures instructions. The PCR product was cleaned up once using SPRIselect reagent (Beckman Coulter B23319) using the manufacturers protocol. DNA was eluted in 18 uL and a first PCR using a i5 adapter forward primer and the ROS1 reverse primer pool was performed. Each PCR contained 10 uL DNA, 25 uL Platinum SuperFi 2× Master Mix (Invitrogen), 2.5 ul of the i5 adapter forward primer and 2.5 uL of the ROS1 reverse primer pool. Cycling was followed using manufactures ins...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Digital information | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com