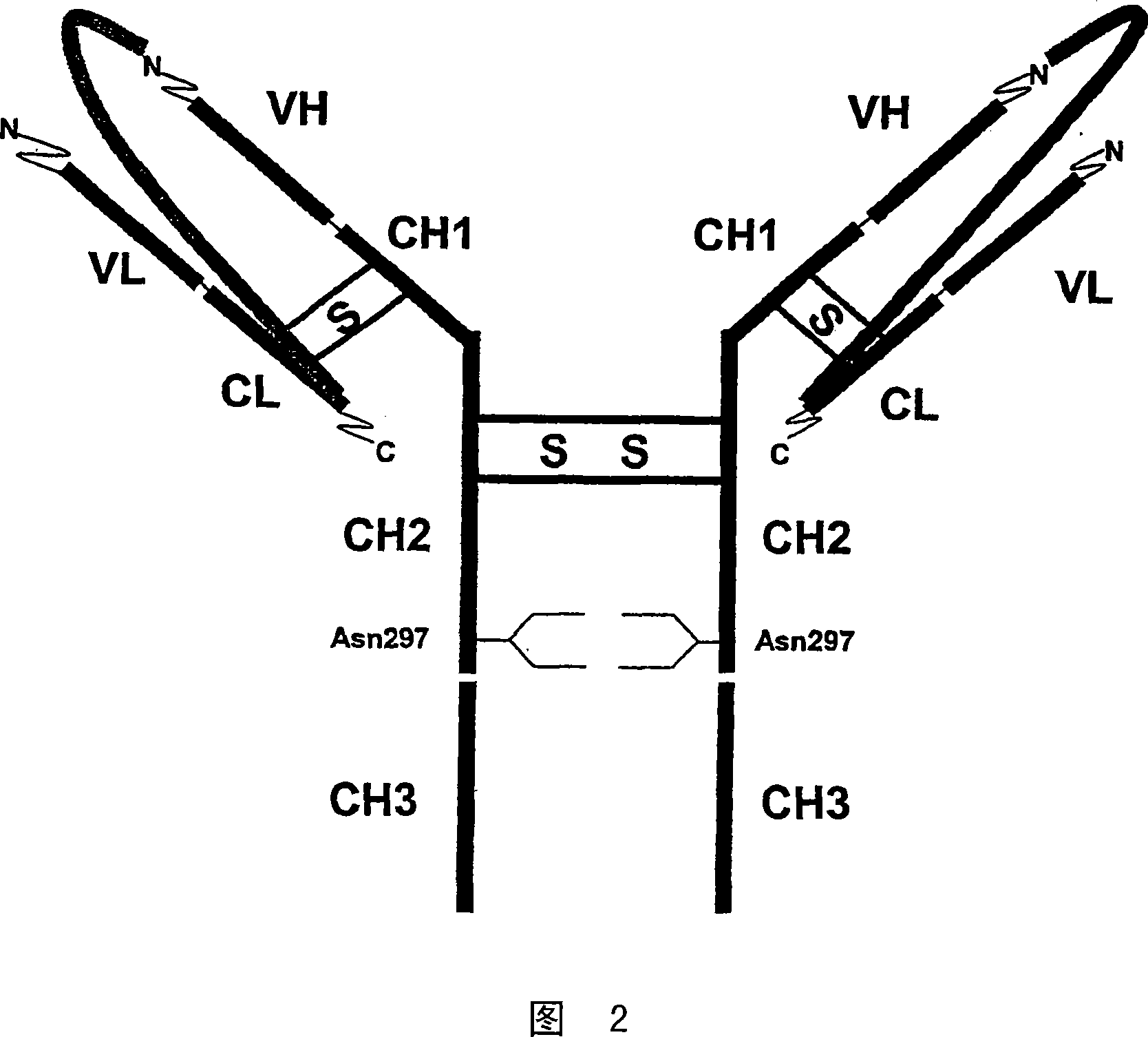
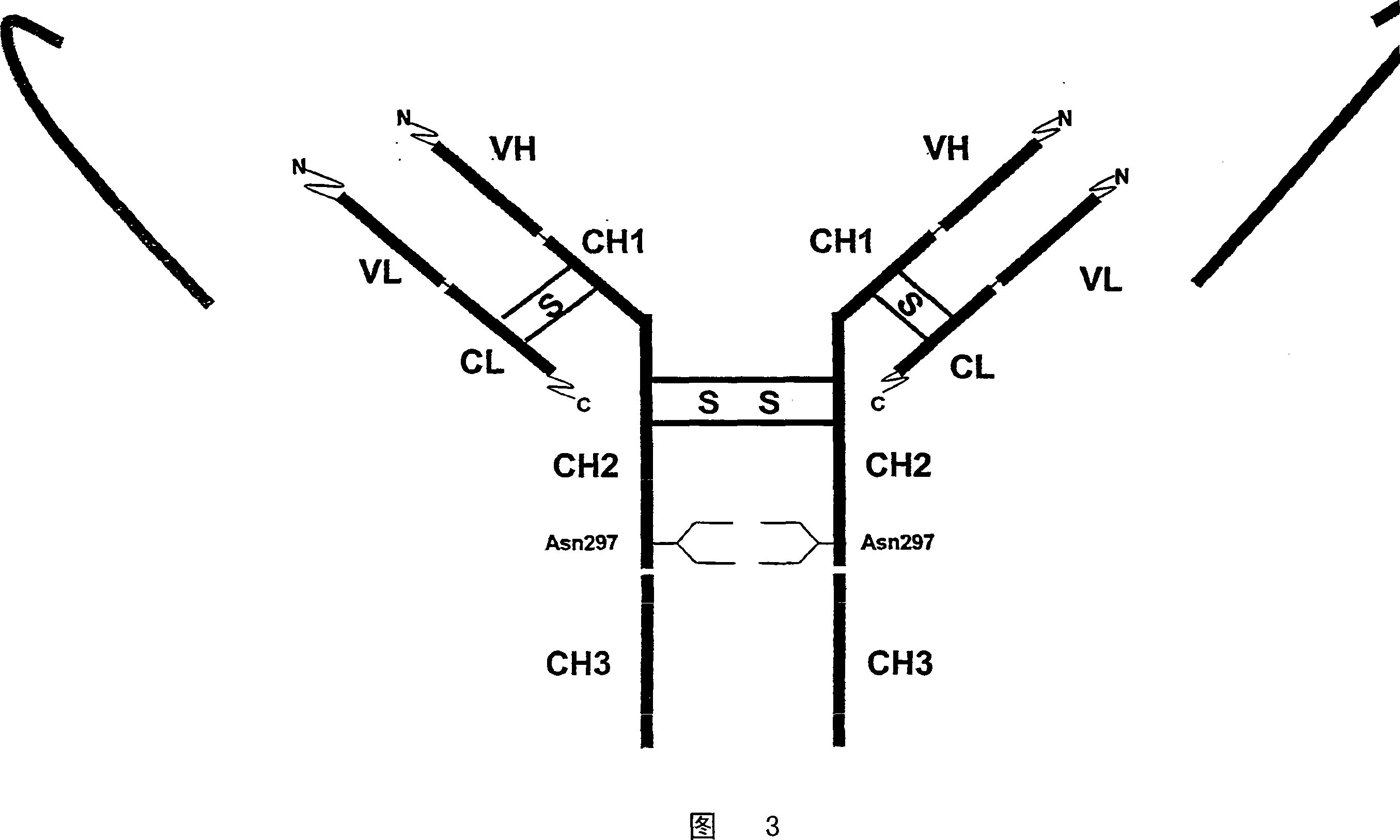
Single chain antibody with cleavable linker
An antibody, proteolytic cleavage technology, applied in the direction of antibodies, antibody mimetics/scaffolds, polypeptides containing positioning/targeting motifs, etc., can solve the problems of lack of constant regions, lack of
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
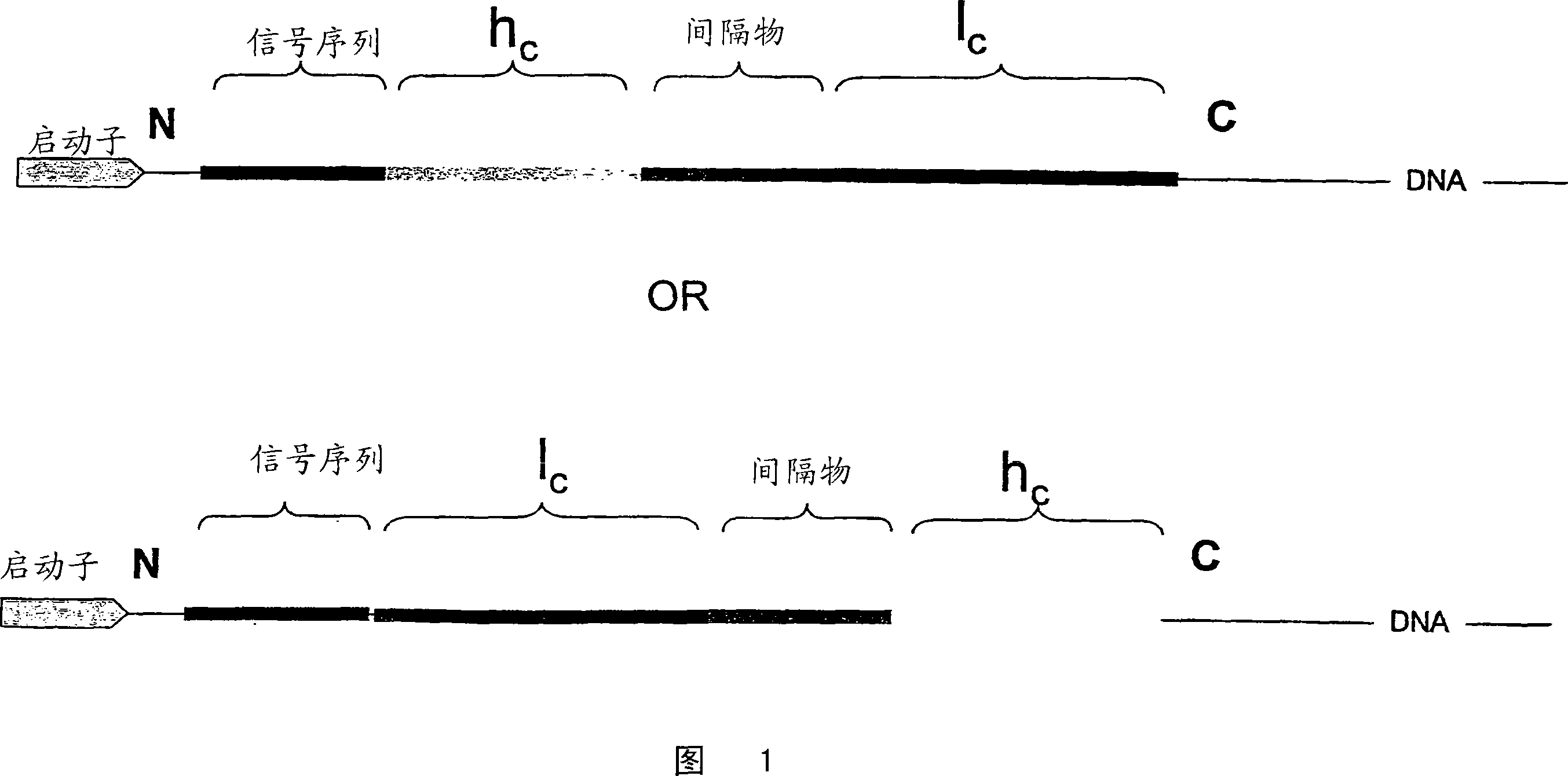
[0102] The fusion protein design for expressing antibody anti-DX is as follows:
[0103] The α-amylase signal sequence is in italics. The spacer peptide between the mature light and heavy chains is underlined. The DNA sequence encoding the signal sequence is:
[0104] The DNA sequence encoding the linker is
[0105] The DNA encoding the light chain variable region of the anti-DX antibody was synthesized by PCR overlap and an Mly1 site was added upstream of the first immunoglobulin chain. DNA encoding the light chain constant region of IgG1 was ordered from GeneArt Inc. DNA encoding the entire light chain was prepared by PCR overlap extension and cloned into the pCR2.1 topo vector. The entire light chain has an Mly1 site at the 5' end and a Not1 site 3' to the stop codon. The DNA encoding the entire light chain was ligated with the DNA encoding the alpha-amylase signal sequence into the pPICZA vector. The α-amylase signal sequence is synthesized from overlapping oligonucl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com