Nucleic acid amplification method
A nucleic acid and oligonucleotide technology, applied in the field of nucleic acid amplification, can solve problems such as long reaction time, troublesome temperature control, and increased time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[0109] The second embodiment of the present invention comprises the following steps: (a) annealing one of the present at least two primers to a nucleic acid strand serving as a template, through the action of the above-mentioned strand displacement type DNA polymerase, from The 3' end of the primer starts to undergo a synthesis reaction, thereby synthesizing an extension product; (b) applying a higher temperature than that in step (a) to the double-stranded nucleic acid obtained by the above-mentioned process without causing the above-mentioned double-stranded In the case of denaturation, the same oligonucleotide primer used in the step (a) is used to infect the double strands, and the synthesis reaction is carried out from the 3' end of the primer by the action of the above-mentioned strand-displacing DNA polymerase , thereby synthesizing an extension product; (c) annealing a primer different from the primer used in the step (a) to the nucleic acid dissociated in the step (b) ...
Embodiment 1
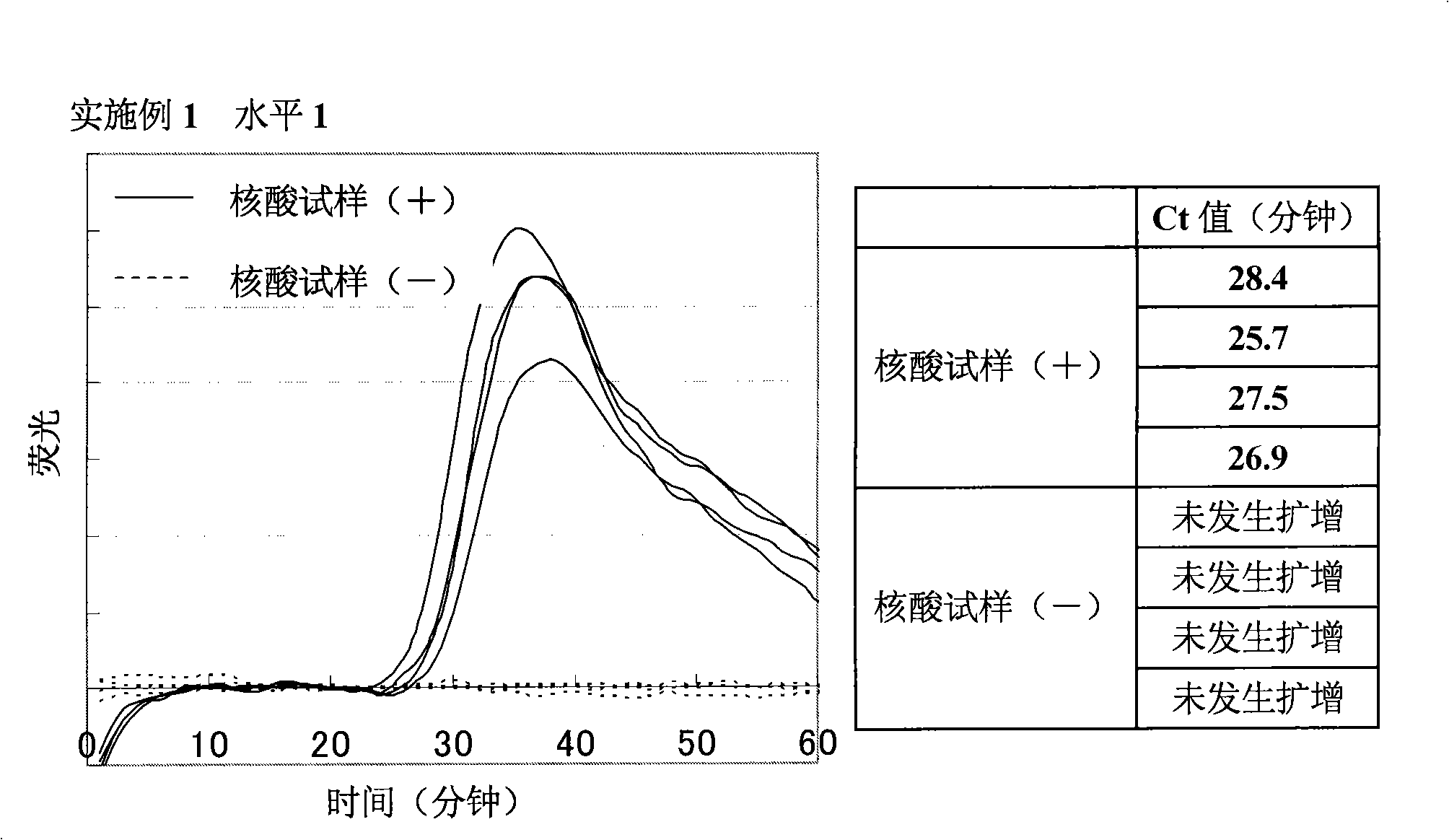
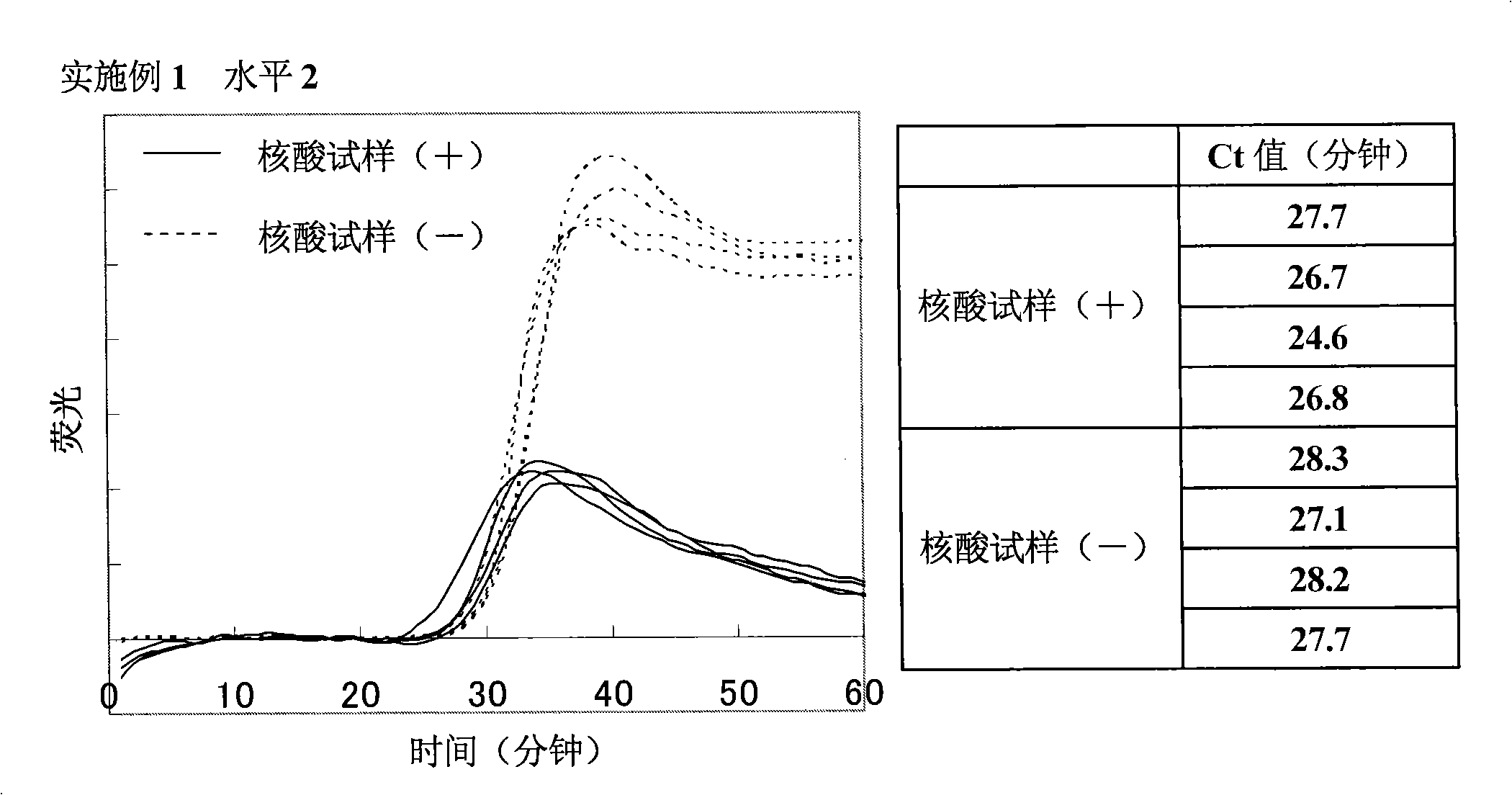
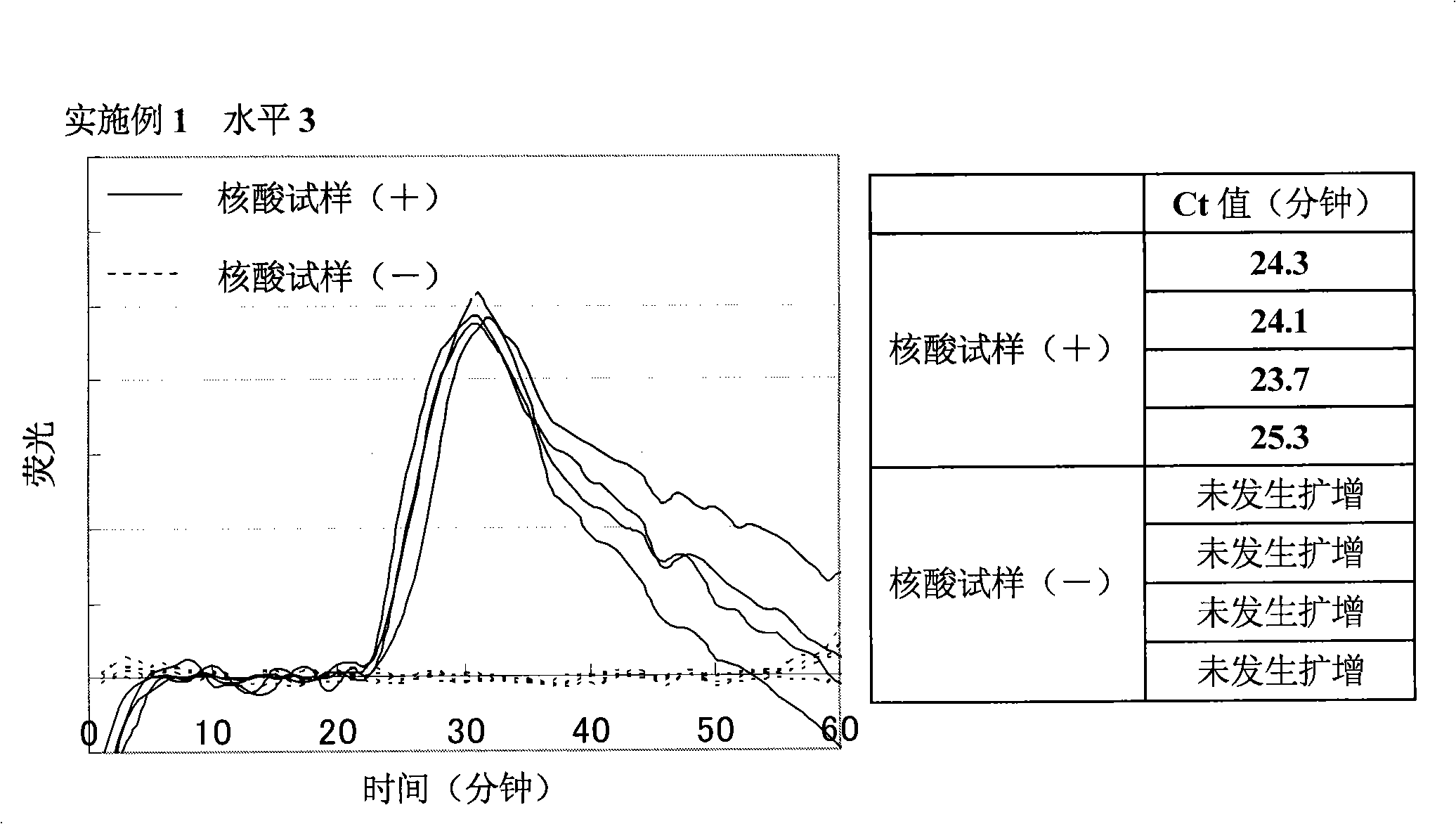
[0167] Amplification of target nucleic acid sequence in human gene
[0168] (1) Preparation of nucleic acid sample solution containing target nucleic acid fragments
[0169] 7.5 ng of Human Genomic DNA (manufactured by Clontech) was heated at 98° C. for 3 minutes, and a specific sequence in the target nucleic acid was amplified under the following conditions. In addition, as a negative control, a sample was prepared by heating purified water under the same conditions as above.
[0170]
[0171] As primers, the following four primers were designed and purchased from Opelon Corporation. The sequences of the respective primers are shown below. Also, primers (1) and (2) are complementary to sequences in the β-actin gene, and primers (3) and (4) are complementary to sequences in the β2AR gene.
[0172] Primer (1):
[0173] 5'-GGGCATGGGTCAGAAGGATT-3' (SEQ ID NO: 1)
[0174] Primer (2):
[0175] 5'-CCTCGTCGCCCACATAG-3' (SEQ ID NO: 2)
[0176] Primer (3):
[0177] 5'-CTTGCT...
Embodiment 2
[0225] Influence of the concentration of surfactant
[0226] (1) Preparation of nucleic acid sample solution containing target nucleic acid fragments
[0227] 7.5 ng of Human Genomic DNA (manufactured by Clontech) was heated at 98° C. for 3 minutes, and a specific sequence in the target nucleic acid was amplified under the following conditions. In addition, as a negative control, a sample was prepared by heating purified water under the same conditions as above.
[0228]
[0229] As primers, primer (1) and primer (2) used in Example 1 were used.
[0230] Primer (1):
[0231] 5'-GGGCATGGGTCAGAAGGATT-3' (SEQ ID NO: 1)
[0232] Primer (2):
[0233] 5'-CCTCGTCGCCCACATAG-3' (SEQ ID NO: 2)
[0234]
[0235] Tween 20 (Formula 1) was used as surfactant.
[0236] (2) Nucleic acid amplification reaction
[0237] Using a reaction solution having the composition shown below, the amplification reaction was carried out at 60°C for 60 minutes. The enzyme used was Bst. Polymerase...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com