Treatment and prevention of influenza
A technology for influenza A virus and nucleic acid constructs, which is applied in the field of nucleic acid constructs and nucleic acid constructs encoding double-stranded RNA molecules, and can solve problems such as high mortality and high pathogenic effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0234] Example 1. Selection of shRNA sequences for inclusion in transgenes
[0235] The most highly conserved genes PB1, PB2, PA, NP and M1 of influenza A were selected for RNAi targeting. The inventors used siVirus (a web-based antiviral siRNA design software for highly divergent viral sequences, Naito et al., 2006) to identify highly conserved regions within selected genes and predict siRNAs capable of screening for shRNA selection sequence. The software highlights some regions within the analyzed influenza A genome segments that are of particular interest for shRNA design, namely the 3' regions of PB1, PB2, PA and NP. More specifically, these are segment 1 (PB2 gene) nucleotides 2240-2341; segment 2 (PB1 gene) nucleotides 2257-2341; segment 3 (PA gene) nucleotides 2087-2233; segment 5 (NP gene) nucleotides 1484-1565.
[0236] The inventors selected 29 siRNA sequences predicted from siVirus to screen for shRNA selection (Table 1). Several algorithms are available for s...
Embodiment 2
[0245] Example 2. Chicken promoter sequence
[0246] It is known that when designing transgenic constructs it is desirable to include a promoter that includes upstream sequences to allow efficient attachment of a polymerase to the promoter sequence. Nonetheless, the chicken U6 promoter was designed and tested to contain the minimum amount of promoter sequence required to cause shRNA transcription, thus allowing for a reduction in the overall size of the transgene construct.
[0247]Two versions of the construct, pcU6-4shNP-1496 and pcU6-4(+100)shNP-1496, were tested for shRNA expression by RNAse protection assay, or for viral silencing. The first plasmid contains the minimal chicken U6-4 sequence required to express the shNP-1496 short hairpin RNA. The second plasmid contained 100 bp of additional cU6-4 promoter upstream sequence. It is expected that the second construct containing 100 bp of additional upstream sequence will provide better shRNA expression.
[0248] Table...
Embodiment 3
[0252] Example 3. Construction of ddRNAi plasmids for expressing selected shRNAs
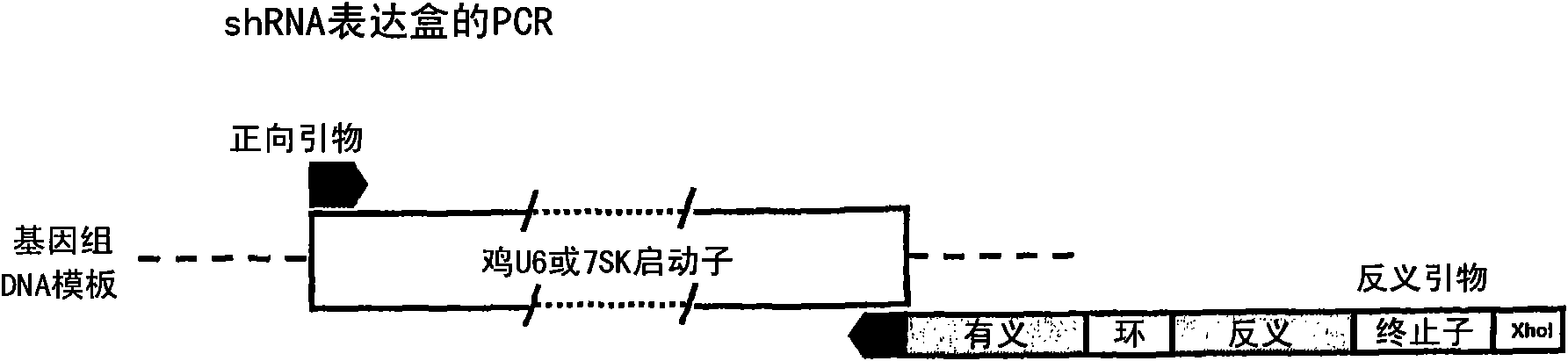
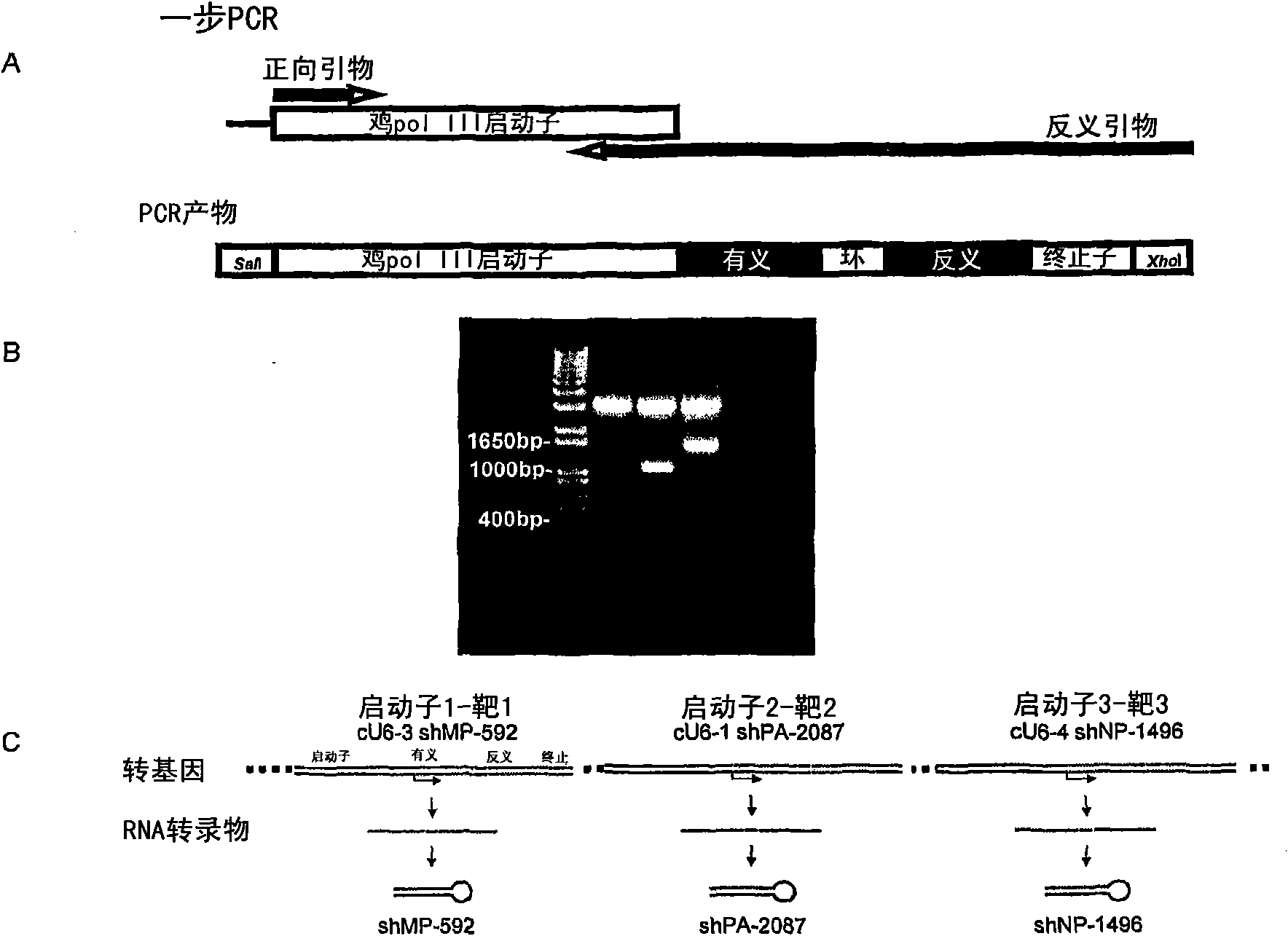
[0253] Chicken polymerase III promoters cU6-1 (GenBank accession number DQ531567), cU6-3 (DQ531569), cU6-4 (DQ531570) and c7SK (EF488955) were used as templates to construct ddRNAi expression of selected shRNAs via one-step PCR Plasmid ( figure 1 ). The PCR for constructing the plasmid uses primers TD135 paired with TD218 or TD275 for the cU6-1 promoter; TD175 paired with TD216, TD274 or TD302 for the cU6-4 promoter; TD176 paired with TD217 for the cU6-3 promoter; TD269 was paired with TD307 or TD316 for the c7SK promoter (see Table 4 for details of primer sequences and specific shRNAs amplified). Reverse primers in each PCR were designed to contain the last 20 nucleotides of each promoter sequence, shRNA sense, loop, and shRNA reaction sequences, and were purified by HPLC. The full-length amplified expression cassette product was ligated into pGEM-T Easy and then sequenced.
[0254] Among...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com