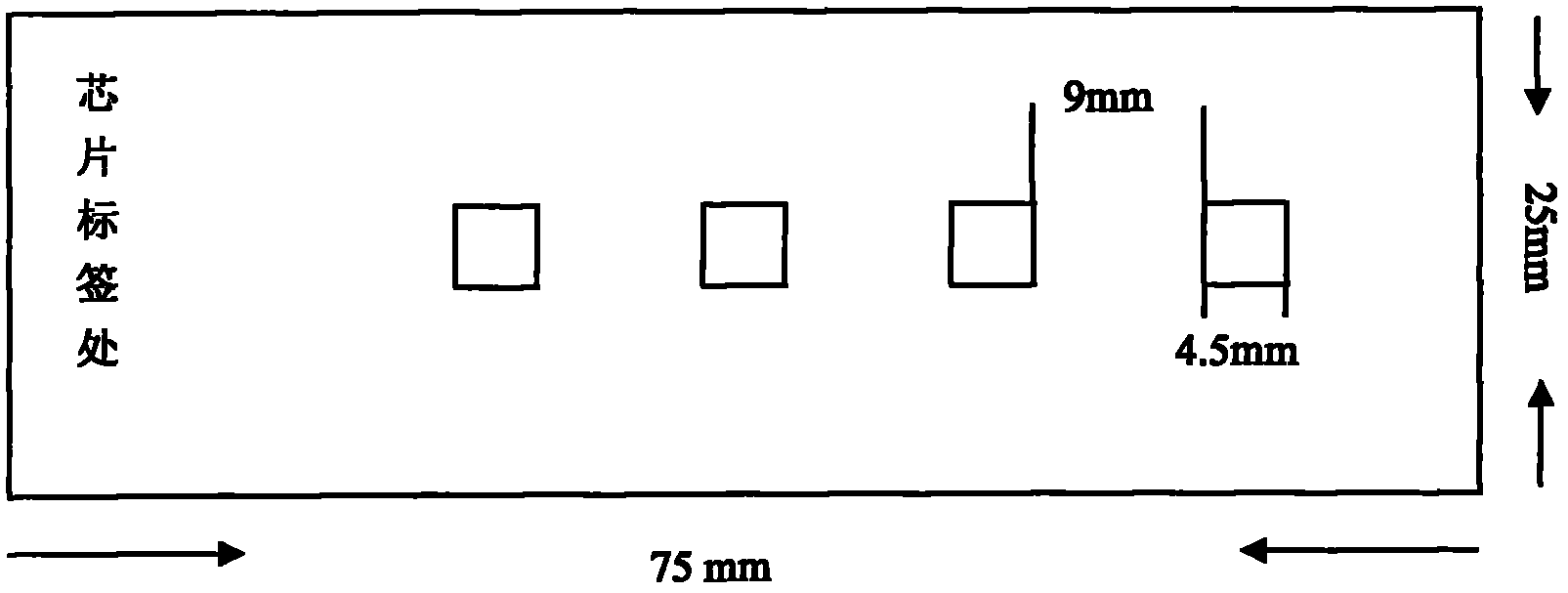
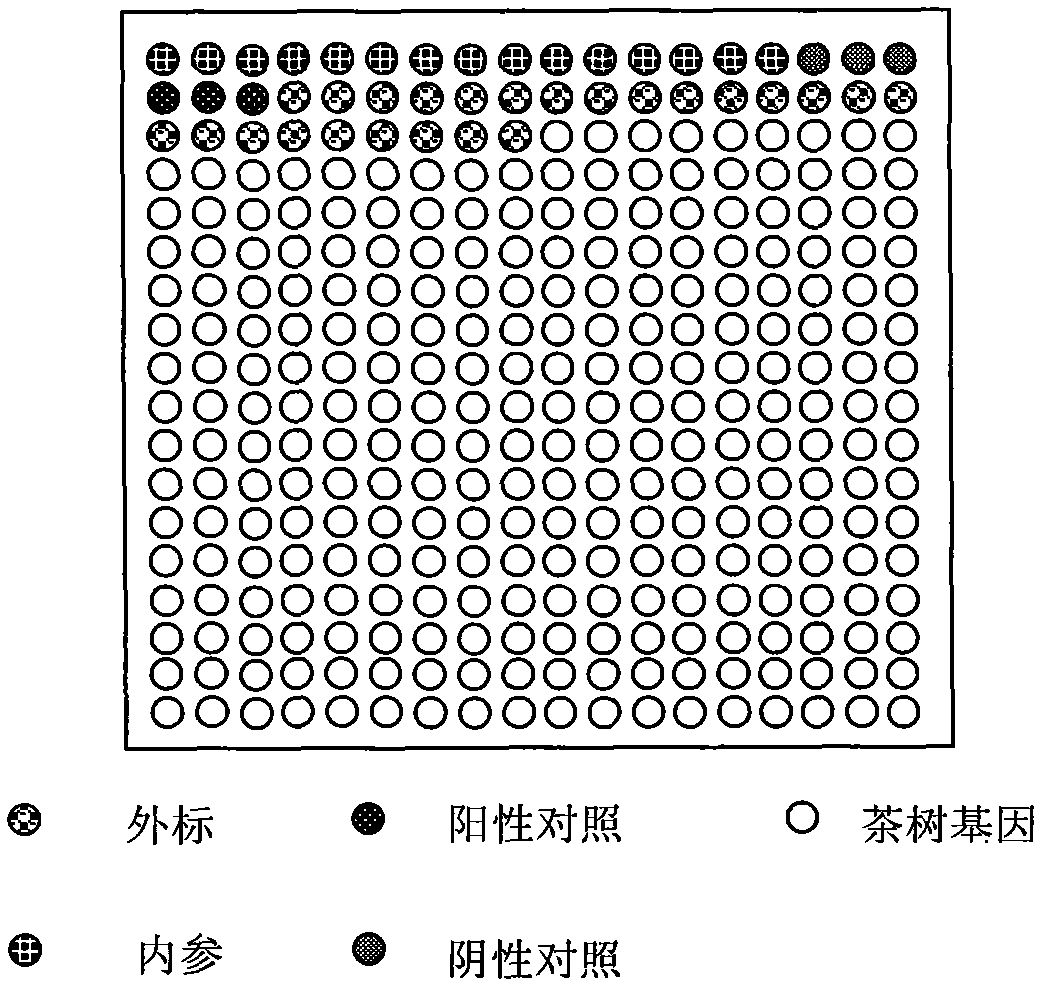
cDNA chip for detecting expression information of genes related to secondary metabolism of tea trees
A technology for secondary metabolism and gene expression, which is applied in the field of cDNA chips for detecting gene expression information related to secondary metabolism of tea trees, and can solve the problem of not being able to analyze genes at the same time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0025] Embodiment 1, the construction of tea tree leaf cDNA library
[0026] 1. Extraction of total RNA from tea tree leaves
[0027] In spring, take 100 mg of vigorously growing young leaves of tea tree, quickly add liquid nitrogen to freeze and grind into a fine powder, add 1000 μL Trizol reagent (purchased from Gibco BRL) to continue grinding, and then add 1000 μL Trizol reagent, mix evenly and divide into two vials. Into a 1.5mL centrifuge tube treated with diethyl pyrocarbonate, place on ice for 5 minutes, add 200 μL of chloroform each, mix by inverting, centrifuge at 12,000 rpm at 4°C for 10 minutes, take 500 μL of the supernatant, add an equal volume of iso Propanol, gently upside down 10 times, place at room temperature for 10 minutes, centrifuge at 12,000 rpm at 4°C for 10 minutes, remove the supernatant, add 1 mL of 75% ethanol to the precipitate to wash gently, pour off the ethanol, and add 20 μL of coke after drying. Diethyl carbonate-treated double distilled wate...
Embodiment 2
[0082] Embodiment 2, the construction of tea tree radicle cDNA library
[0083] 1. Extraction of total RNA from young roots
[0084] Sow the stored seeds of the tea tree Longjing 43 in quartz sand with a certain humidity, and cultivate them in a culture room at room temperature of 25°C. After the seedlings grow, take 100 mg of their young roots, quickly add liquid nitrogen to freeze and grind them into fine powder, and add 1000 μL Trizol After the reagent continues to be ground, add 1000 μL of Trizol reagent, mix evenly and divide into two 1.5mL centrifuge tubes treated with DEPC (diethyl pyrocarbonate), place on ice for 5 minutes, add 200 μL of chloroform each, mix upside down, Centrifuge at 12,000 rpm at 4°C for 10 minutes, take 500 μL of supernatant, add an equal volume of isopropanol, invert up and down gently 10 times, place at room temperature for 10 minutes, centrifuge at 12,000 rpm at 4°C for 10 minutes, remove the supernatant, add 1 mL of 75% ethanol to the precipitat...
Embodiment 3
[0152] Example 3, Sequencing of Tea Tree Leaf and Young Root Expressed Sequence Tags
[0153] Using the previously constructed tea tree leaf cDNA library and tea tree radicle cDNA library as the source of sequencing EST libraries, the automatic sequencer Megabace TM 1000 Random sequencing analysis was performed on 4320 clones of the Longjing 43 shoot cDNA library, and 2963 effective sequences were obtained. After deduplication and splicing of all effective sequences, 1692 shoot expression sequence tags were obtained; 5115 of the radicle cDNA library The clone was subjected to random sequencing analysis, and 4833 effective sequences were obtained, most of which were between 300bp and 700bp in length, with an average length of 474bp. After deduplication and splicing of all effective sequences, 3482 radicle expression sequence tags were obtained. All the obtained non-redundant sequences were compared with the protein database of GenBank by BlastX analysis, and the results were m...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap