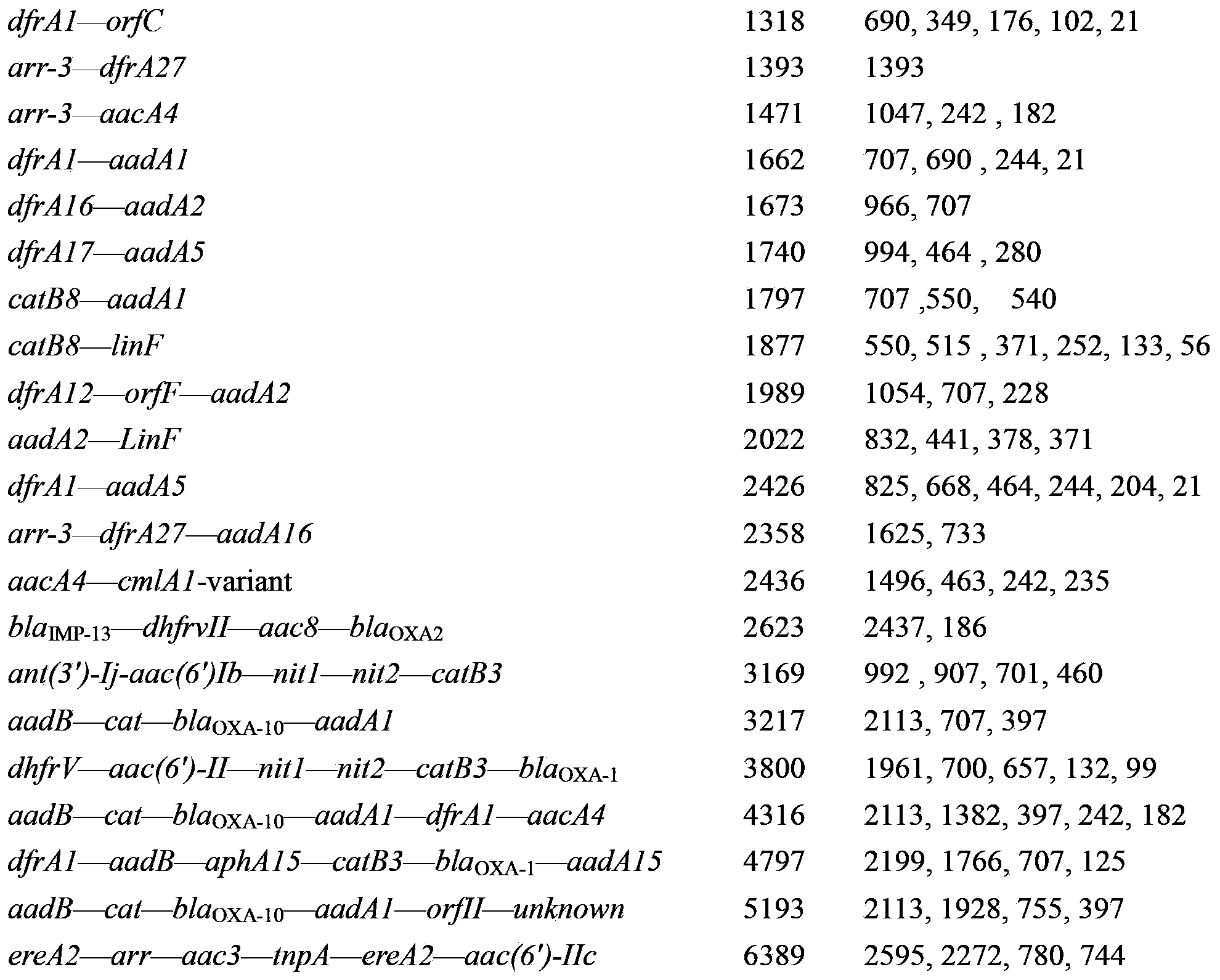
Method for detecting gene cassette array in bacterium integron by using EcoRII enzyme digestion restriction map library
A technology of integrons and gene cassettes, applied in the field of rapid detection of gene cassette arrays in bacterial integrons, can solve the problems of high experimental costs and long experimental processes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1. Screening of drug-resistant bacteria
[0039] 1. Take the wastewater samples around the hospital and quickly put them in the medical wide-mouth sampling bottles after high-pressure steam sterilization. The general sampling volume is about 500ml. Put the collected water samples in the ice box and bring them back to the laboratory within 4 hours. filter processing.
[0040] 2. If the sample water sample has a lot of dirt, you can use sterile qualitative filter paper to perform coarse suction filtration to remove more impurities, so as to avoid extremely slow processing speed with sterile filter membrane.
[0041] 3. Pass the coarsely filtered sample water through a sterile sand core filter device 0.22μm sterile filter membrane) for filtration, the bacteria were enriched on the membrane, and the filter membrane was transferred to 100mL LB liquid medium (5g of yeast extract, 10g of tryptone, 10g of NaCl, dissolved in 800ml of distilled water, and adjusted to pH...
Embodiment 2
[0048] Example 2. Extract 247 drug-resistant bacterial genomic DNAs with the CTAB method
[0049] 1. Bacteria culture: Inoculate 247 test bacteria in LB liquid medium, shake and culture at 37°C for 16-18 hours to obtain enough bacteria.
[0050] 2. Take 1.5ml of bacterial liquid and centrifuge at 8000rmp / min for 2min in EP tube to collect the bacterial cells.
[0051] 3. Add 500ulTE resuspended bacteria (instrument: vortex shaker) and centrifuge at 8000rpm / min for 2min.
[0052] 4. Remove the supernatant, pay attention to blot the excess water, add 680ulTE buffer solution to the sediment, pipette the resuspended bacteria repeatedly, mix well, add 36ul10% SDS, mix well to avoid the degradation of DNA by nuclease and mechanical oscillation, Add proteinase K on ice, and incubate at 37 °C for more than 1 h until the solution becomes clear. After clarification, add 120ul 5mol / l NaCl, mix well, then add 96ul preheated CTAB / NaCl solution, after mixing, warm bath at 65°C for 10min, ...
Embodiment 3
[0059] Example 3. Screening type 1 integron strains from drug-resistant bacteria
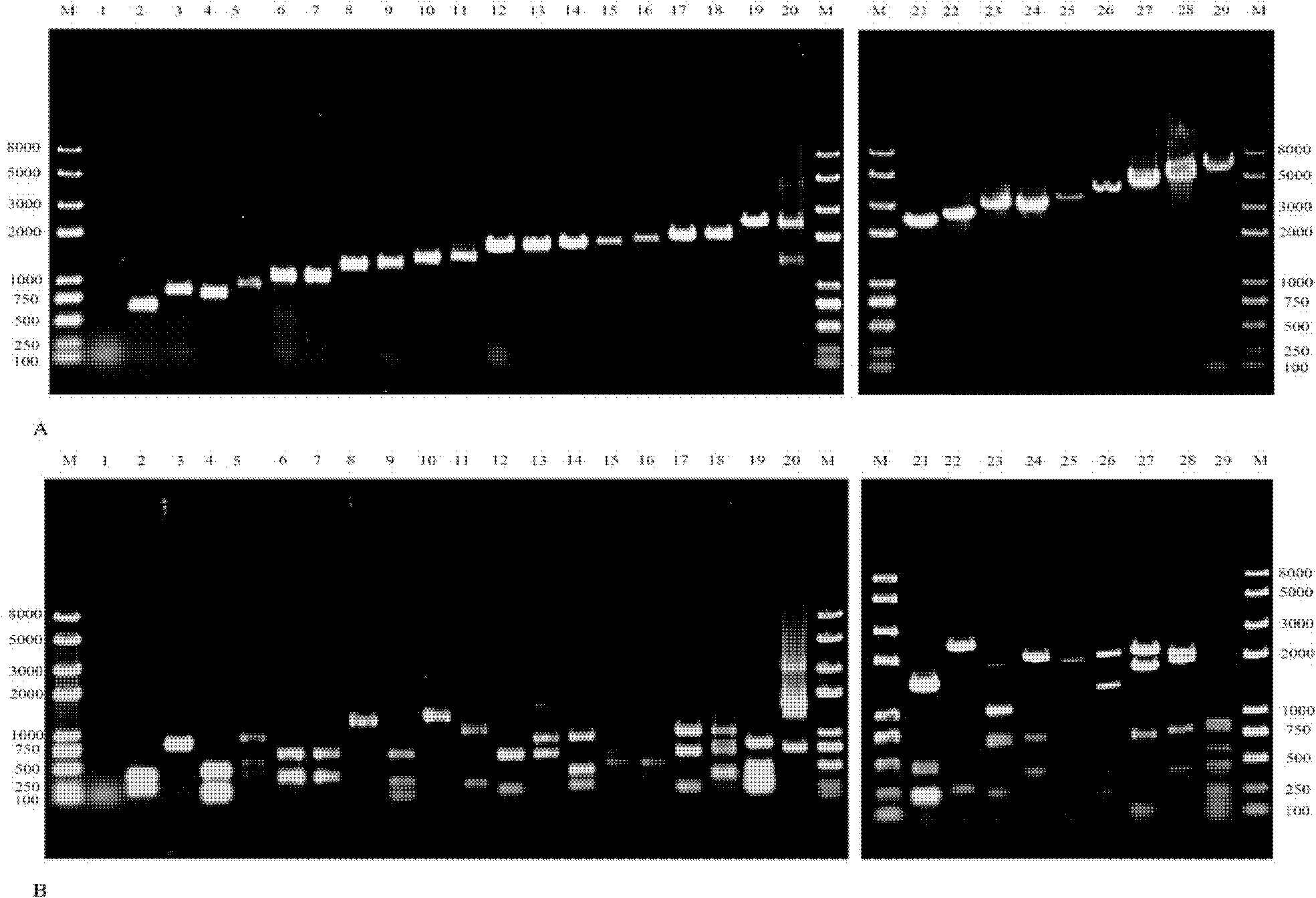
[0060]The genomic DNA of the screened 247 drug-resistant bacteria was used as templates for PCR amplification, and the amplified products were detected by agarose gel electrophoresis to determine whether the drug-resistant bacteria contained type 1 integrase gene. Whether the strain contains type 1 integron. Wherein the amplification primers used for the above PCR are:
[0061] Upstream primer intI1-F: 5'-GTTCGGTCAAGGTTCTGG-3'&
[0062] Downstream primer intI1-R: 5'-CGTAGAGACGTCGGAATG-3'.
[0063] The reaction system and reaction procedure of PCR are as follows:
[0064]
[0065] Test results: Among the 247 strains of drug-resistant bacteria, 146 strains were tested positive for integrase, which means they were drug-resistant bacteria containing type 1 integrons.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com