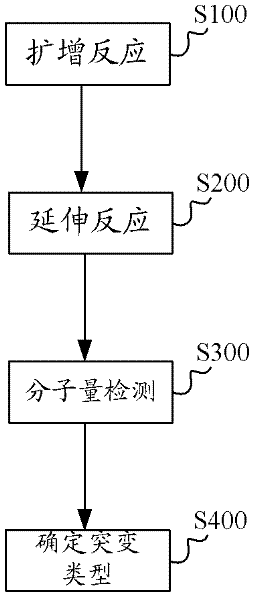
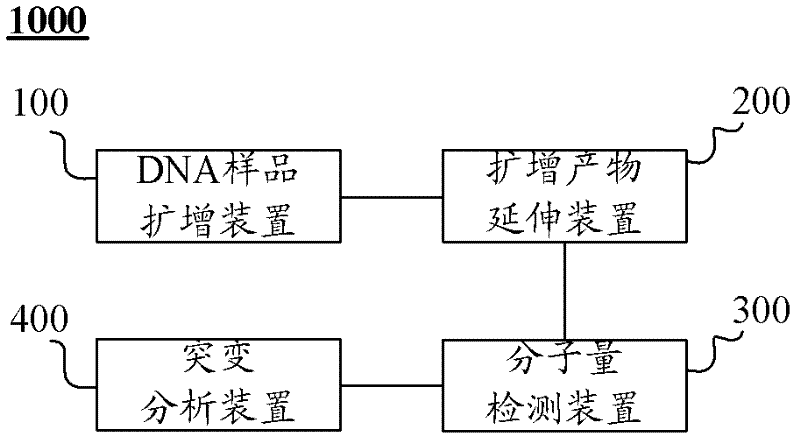
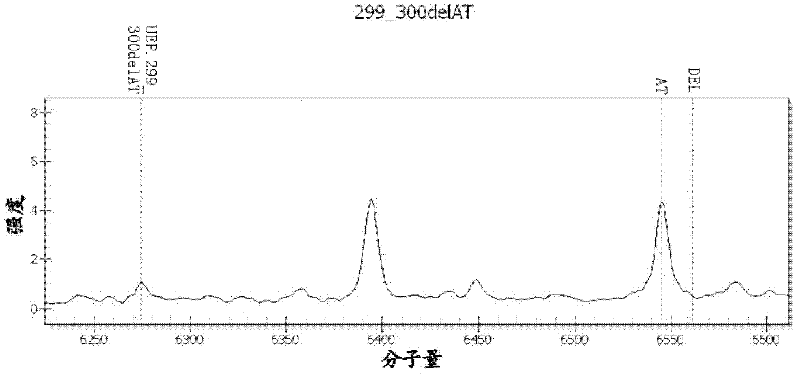
Kit, method and application for detecting mutation of predetermined locus in DNA sample
A technology for pre-determining sites and samples, which is applied in recombinant DNA technology, DNA/RNA fragments, biochemical equipment and methods, etc., and can solve problems such as specific site mutations that need to be improved.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0106] ①Primer design
[0107] According to the selected 20 deafness gene mutation sites to be detected and / or typed, including 4 genes, they are: GJB2 gene (including sites 35delG, 167delT, 176-191del16, 299_300delAT and 235delC), GJB3 gene ( Including sites 538C>T and 547G>A), SLC26A4 gene (including sites 281C>T, 589G>A, 919-2A>G, 1174A>T, 1226G>A, 1229C>T, 1975G>C, 2027T> A, 2162C>T, 2168A>G and IVS15+5G>A) and 1494C>T and 1555A>G of mt DNA.
[0108] Design an amplification primer pair and an extension primer according to the position and genotype of the mutation site, wherein the length of the amplification primer is about 30 bases, and the amplification primer has a tag sequence of 10 bases acgttggatg at the 5′ end; The length of the extension primer is 17-28 bases, allowing 1-5 bases at the 5' end to be incompletely paired with the template, and the bases at the 3' end are completely matched with the adjacent bases at the 3' end of the mutation site. In addition, the ...
Embodiment 2
[0139] ①Primer design
[0140]According to the selected 14 deafness gene mutation sites to be detected and / or typed, they are distributed in 3 genes, namely: GJB2 gene (including sites 299_300delAT and 235delC), SLC26A4 gene (including sites 281C>T , 919-2A>G, 1174A>T, 1226G>A, 1229C>T, 1975G>C, 2027T>A, 2162C>T, 2168A>G and IVS15+5G>A) and 1494C>T and 1555A of mt DNA >G. The amplification primers designed by the present invention for the above 14 mutation sites of deafness genes are selected from the primers in Example 1, see Table 7. The extension primers designed by the present invention for the above 14 mutation sites of deafness genes are selected from the primers in Example 1, see Table 8.
[0141] Table 7: Amplification primers for the above 14 mutation sites of deafness genes.
[0142]
[0143] Table 8: Extension primers for the above 14 mutation sites of deafness genes.
[0144]
[0145]
[0146] Subsequent experimental steps (namely: DNA extraction, PCR ...
Embodiment 3
[0153] ①Primer design
[0154] According to the selected 4 deafness gene mutation sites to be detected and / or typed, they are distributed in 3 genes, namely: GJB2 gene (including site 235delC), SLC26A4 gene (including site 919-2A>G ) and 1494C>T and 1555A>G of mt DNA. Refer to Table 9 for the amplification primers designed by the present invention for the above four mutation sites of deafness genes, and refer to Table 10 for the extension primers.
[0155] Table 9: Amplification primers for the above four mutation sites of deafness genes.
[0156]
[0157] Table 10: Extension primers for the above four mutation sites of deafness genes.
[0158] serial number
Primer sequence
Primer description
SEQ ID NO: 37
ggATCCTGCTATGGGCC
Extension primer for position 235delC
SEQ ID NO: 42
GCAGTAGCAATTATCGTC
Extension primer for position 919-2A>G
SEQ ID NO: 51
CGTACACACCGCCCGTCAC
Extension primer for position 1494C>T
S...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com