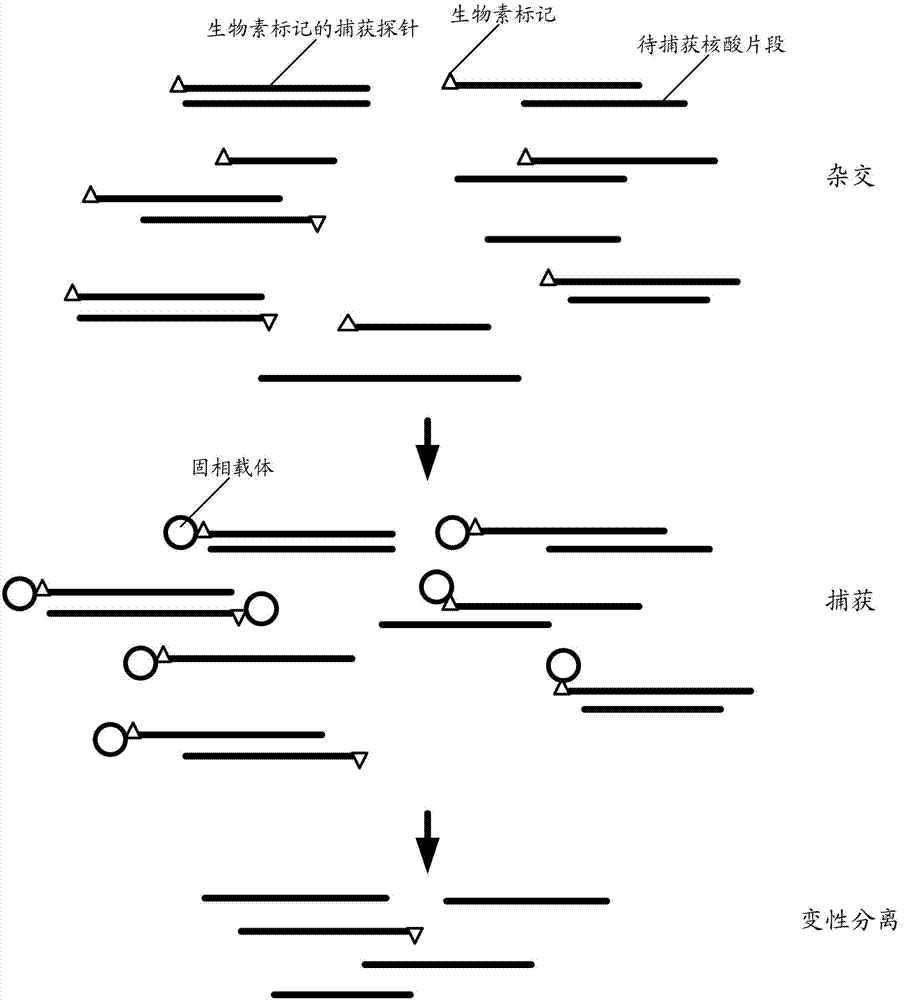
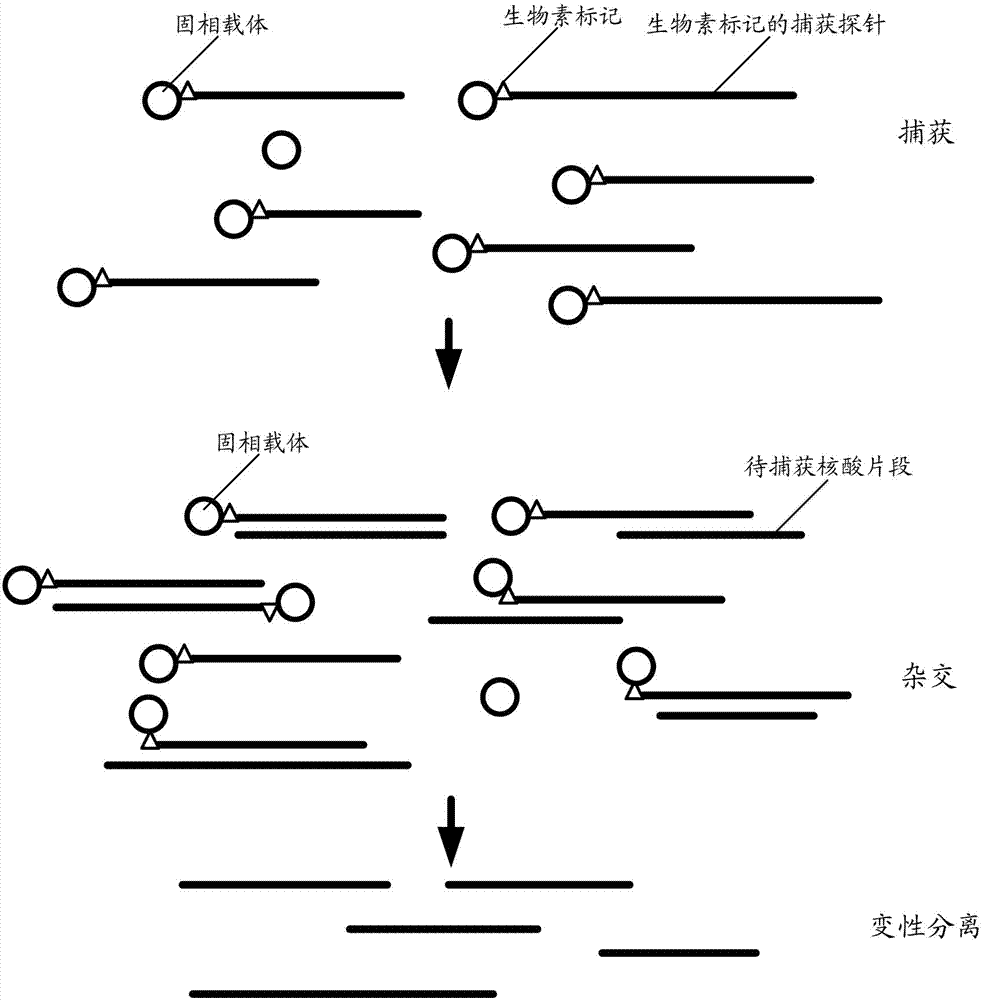
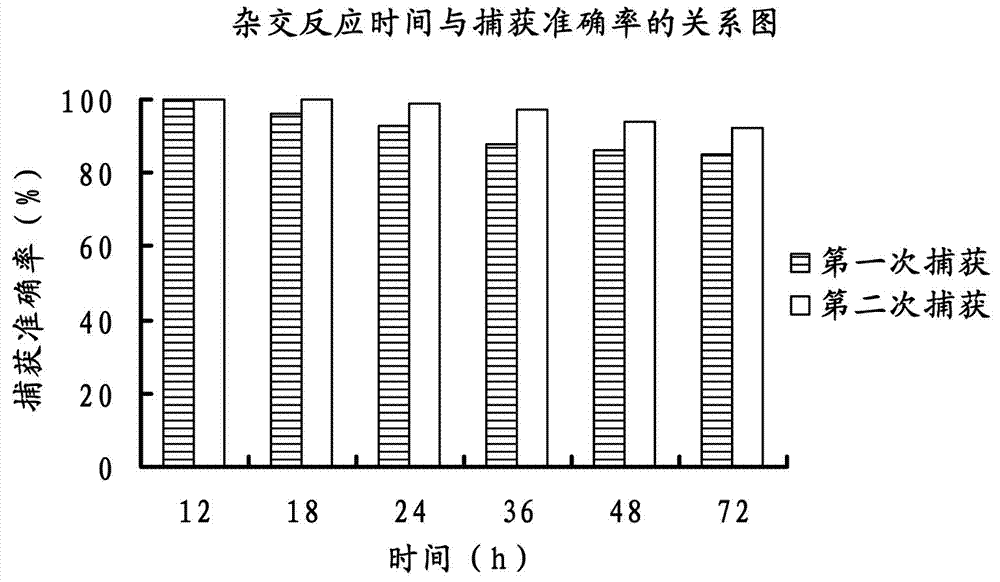
Method for capturing nucleic acid fragment
A technology for capturing nucleic acids and fragments, applied in the fields of genetic engineering and molecular biology, can solve problems such as poor uniformity and poor capture effect, achieve good uniformity, avoid poor capture effect, and reduce production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
no. 1 example
[0061] The present invention proposes a first embodiment, the nucleic acid molecule containing the nucleic acid fragment to be captured is a cloning carrier library, and there are multiple nucleic acid fragments to be captured and constructed into multiple cloning vectors respectively, and the step A includes the following steps:
[0062] A01. Use restriction enzymes to cut off the nucleic acid fragments to be captured in the cloning vector library to obtain DNA templates;
[0063] A02. Fragmenting the DNA template to obtain DNA template fragmentation products;
[0064] A03. The DNA template fragmentation product is connected with a biotin-labeled adapter element to obtain a biotin-labeled capture probe.
[0065] In this protocol, the nucleic acid fragment to be captured is digested from the cloning vector by enzyme digestion, and then connected with a biotin-labeled adapter element to obtain a biotin-labeled capture probe. The cloning vectors in the cloning vector library in...
no. 3 example
[0078] The present invention also proposes a third embodiment, the step A includes the following steps:
[0079] A21. Fragmentation of nucleic acid molecules containing nucleic acid fragments to be captured to obtain fragmentation products;
[0080] A22. The fragmentation product is connected with the adapter element to obtain a nucleic acid fragment containing the adapter;
[0081] A23. Perform PCR amplification on the product of step A22 using biotin-labeled amplification primers to obtain biotin-labeled capture probes.
[0082] It should be noted that the nucleic acid molecule containing the nucleic acid fragment to be captured is a PCR amplification product, and the PCR amplification product uses corresponding PCR primers to amplify the target region on genomic DNA, mitochondrial DNA, plasmid or RNA acquired. The target area is composed of nucleic acid fragments to be captured. There may be multiple target areas. The linker element is a nucleic acid molecule, used for ...
no. 4 example
[0087] The present invention also proposes a fourth embodiment, the step A includes the following steps:
[0088] A31. Fragmentation of nucleic acid molecules containing nucleic acid fragments to be captured to obtain fragmentation products;
[0089]A32. The fragmented product is connected to a biotin-labeled adapter element to obtain a biotin-labeled capture probe.
[0090] It should be noted that the nucleic acid molecule containing the nucleic acid fragment to be captured is a PCR amplification product, and the PCR amplification product uses corresponding PCR primers to amplify the target region on genomic DNA, mitochondrial DNA, plasmid or RNA acquired. The target area is composed of nucleic acid fragments to be captured. There may be multiple target areas. The ligation of the fragmented product with the biotin-labeled adapter element can only occur at one or both ends of the fragmented product of the DNA template.
[0091] The biotin-labeled linker element is a nuclei...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com