Method for detecting cadmium pollution through variation of genetic expression of brassica plant
A technology of gene expression and Brassica genus, applied in the field of molecular biology detection of environmental pollution, achieves the effects of strong sensitivity and specificity, improved identification efficiency, and high practical value
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
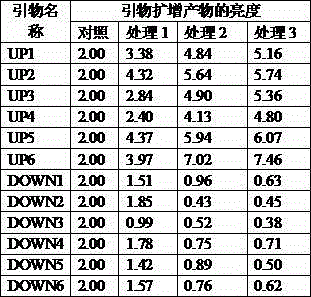
[0032] Identification of up-regulated and down-regulated genes specifically expressed in Brassica juncea under cadmium induction.
[0033] Select plump and clean mustard seeds, soak them in 75% ethanol for 1 min, then sterilize them in 0.1% mercuric chloride for 10 min, wash them with sterile water for 3 times, soak them in sterile water for 5 hours, and blot the excess water with filter paper , inoculated on 1 / 2 MS solid medium with pH 6.0, containing 5g / L sucrose and 0.7% agar. Cadmium (CdCl 2 ) concentration level (as Cd 2+ 0mg / kg, 0.5mg / kg, 1.0mg / kg and 1.5mg / kg, respectively, grown in a greenhouse under sterile conditions for 15 days, the day-to-night ratio was 14 / 10h, and the day-to-night temperature was 22 / 16°C. The light intensity is 200 μmol m –2 the s –1 . Rapeseed root in solid medium was taken for experiment.
[0034]Collect cadmium-treated and control mustard roots, extract total RNA, hybridize and analyze with 29K Arabidopsis expression chips, and perform ...
Embodiment 2
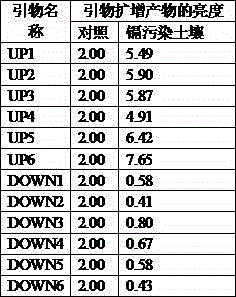
[0050] 1) Take paddy soil from the surrounding farmland of Hunan University of Science and Technology in Xiangtan, Hunan. The content of Pb is 55.4 mg / kg, the content of Zn is 157.8 mg / kg, the content of Cd is 0.17 mg / kg, and the pH value is 6.5.
[0051] 2) Take a flower pot with a diameter of 20cm and a height of 20cm. The following cultures were placed in the flower pots: paddy soil, referred to as control; paddy soil and 0.50 mg / kg Cd, referred to as treatment 1; paddy soil and 1.00 mg / kg Cd, referred to as treatment 2; paddy soil and 1.50 mg / kg Cd, referred to as treatment 2; Referred to as treatment 3. The volume of the culture is about 4 / 5 of the volume of the flower pot. There is a watertight plate under the flower pot. After the pot is installed, water 200ml per pot and place it in the natural environment of Hunan University of Science and Technology for 1 month.
[0052] 3) In September, sow the mustard seeds in the above-mentioned flower pots, place them in the na...
Embodiment 3
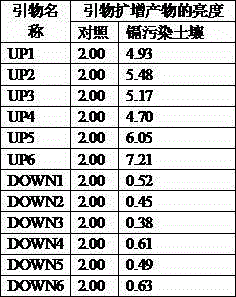
[0060] 1) Collect mustard greens grown in cadmium-contaminated soil and non-polluted soil around a tailing mine in Hunan. Taking non-polluted soil as a control, the cadmium content was determined by atomic absorption spectrometry to be 0.21mg / kg, which is in line with the national soil environmental quality secondary standard (GB15168-1995). The content of cadmium in cadmium-contaminated soil was determined to be 2.23mg / kg by atomic absorption spectrometry. The roots of mustard were collected for extraction of total RNA. The extracted total RNA was reverse transcribed.
[0061] 2) Taking the expression of Actin gene as a reference, the sequence of forward primer ACTF and ACTF is: 5'-GACATTCAACCTCTTGTTTGC-3', and the sequence of reverse primer ACTR is: 5'-CTGCTCGTAGTCAAGAGCAATG-3'; detection by semi-quantitative PCR The expression of 6 all up-regulated genes and 6 all down-regulated genes specifically expressed under the induction of mustard cadmium, the specific operation is...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com