Computer-implemented method and computer system for rank normalization for differential expression analysis of transcriptome sequencing data
A technology for transcriptome sequencing and differential expression, which is used in computing, electrical digital data processing, special data processing applications, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
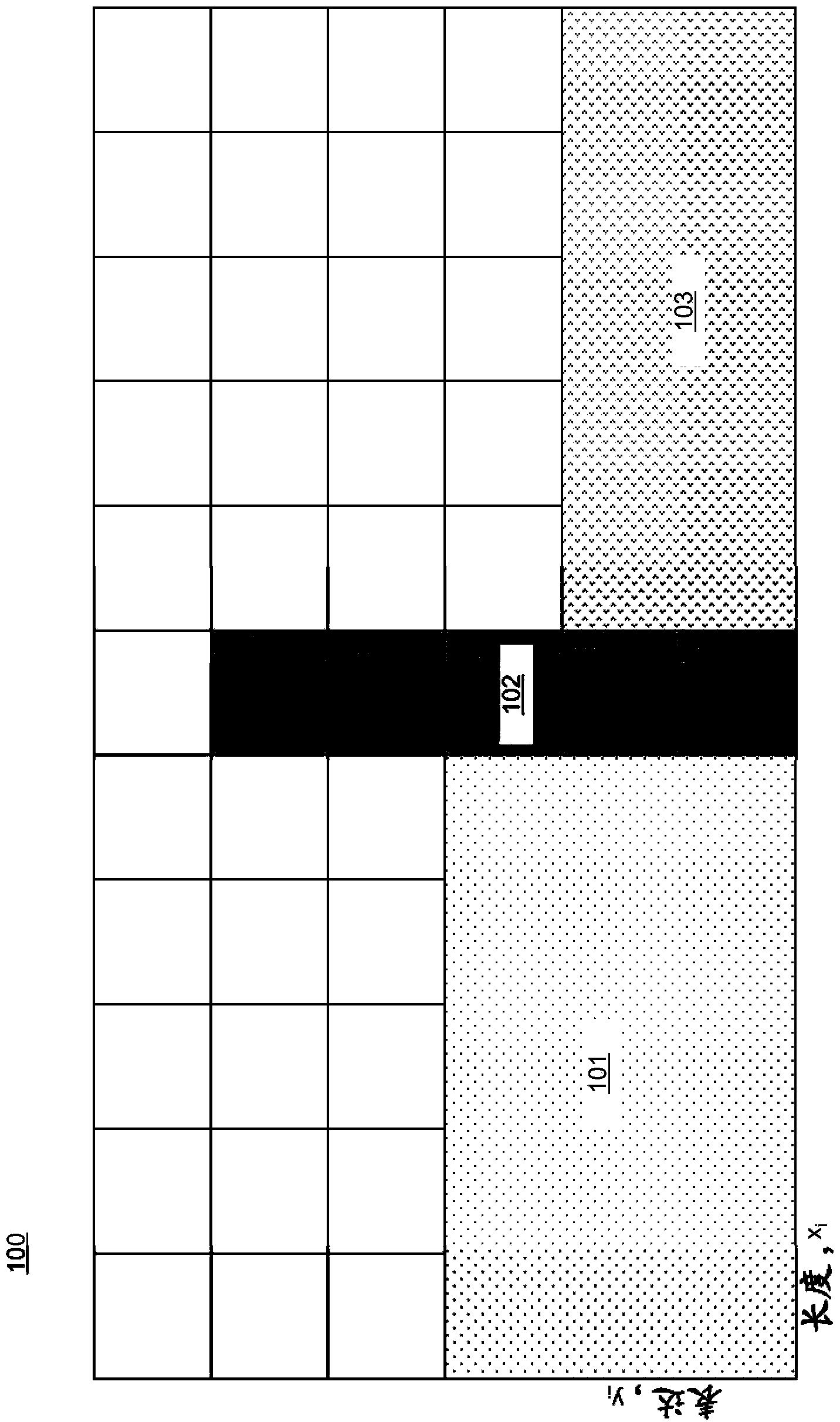
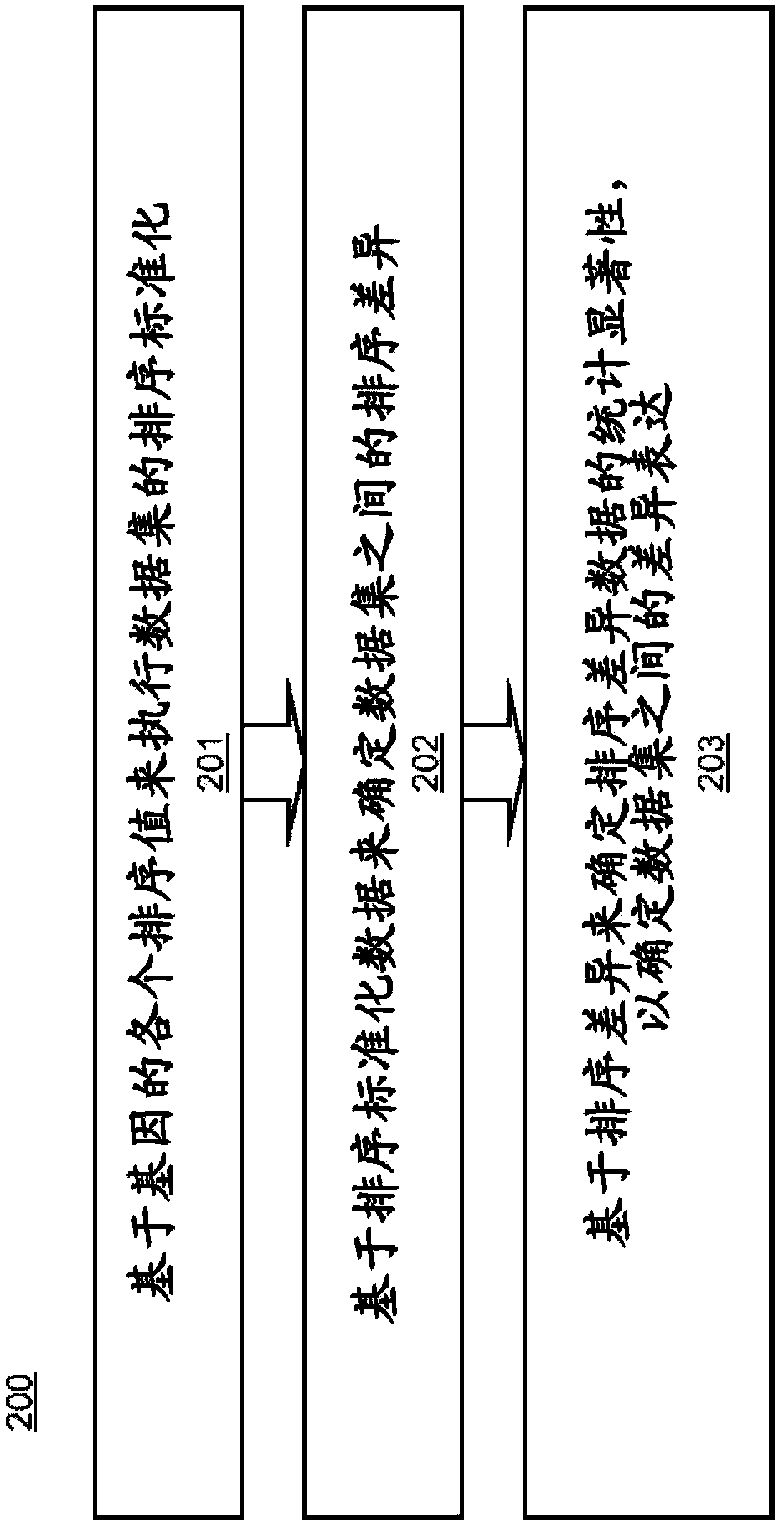
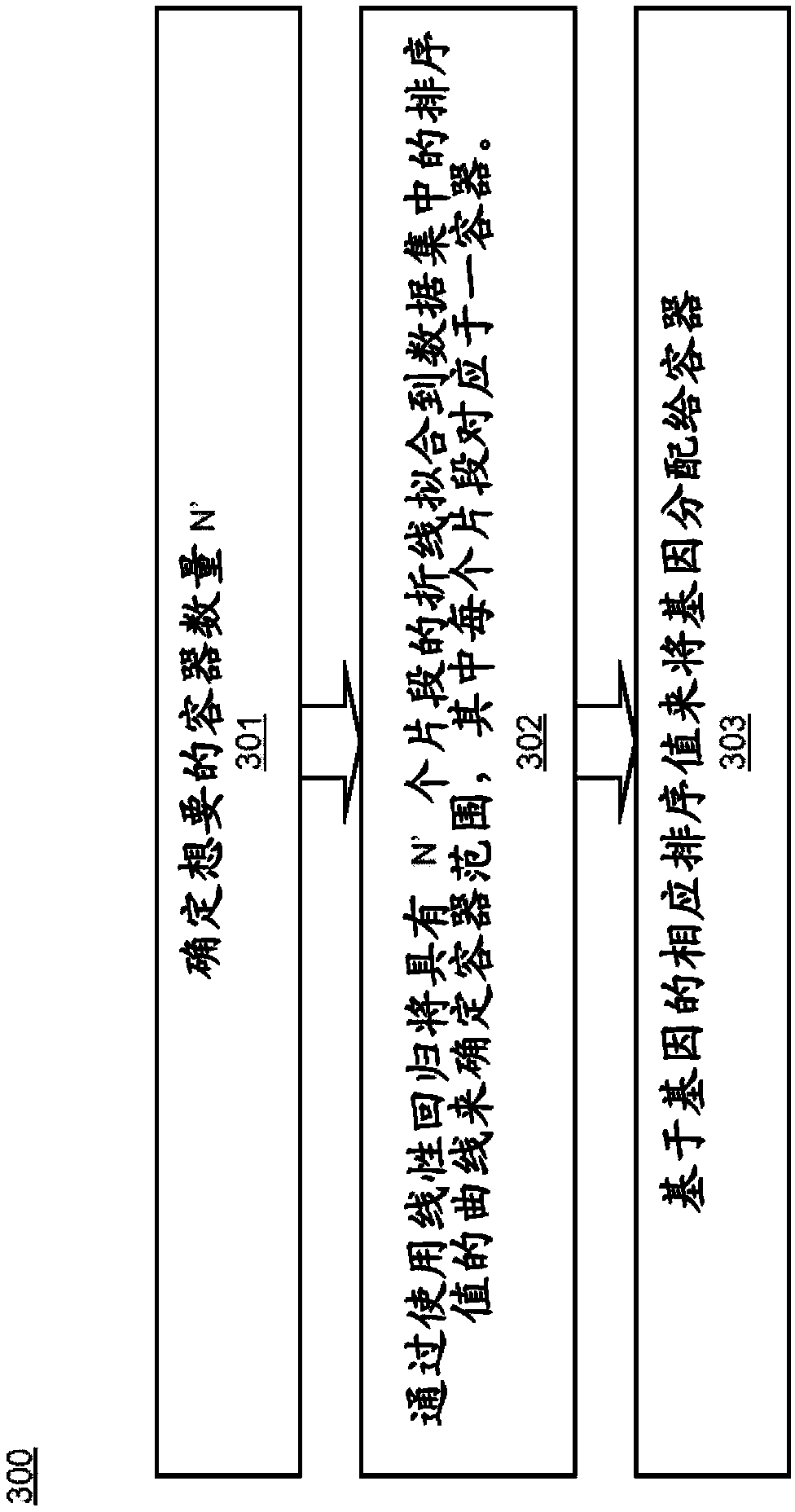
[0014] Embodiments of systems and methods for rank normalization for differential expression analysis of transcriptome sequencing data are provided, and exemplary embodiments are discussed in detail below. Normalization of transcriptome sequencing data can be based on the relative position of genes in the dataset relative to other genes in the dataset. As used herein, the term gene may also refer to any transcriptome sequencing data, including transcripts or mRNA in various embodiments. Ordinal normalization of genetic data yields a unitless number for each gene that can be used for comparison between datasets. Rankings can be determined for individual genes in a dataset, and then gene-specific ranking differences can be determined between datasets. The two data sets being compared may include transcriptome sequencing data from two different samples in some embodiments, or may include transcriptome sequencing data from a single sample at two different time points in other emb...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com