Methods and related devices for single molecule whole genome analysis
A base, part of the technology, applied in the field of nanotechnology and single-molecule genome analysis, can solve problems such as isolation, incompatibility, and cumbersome
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
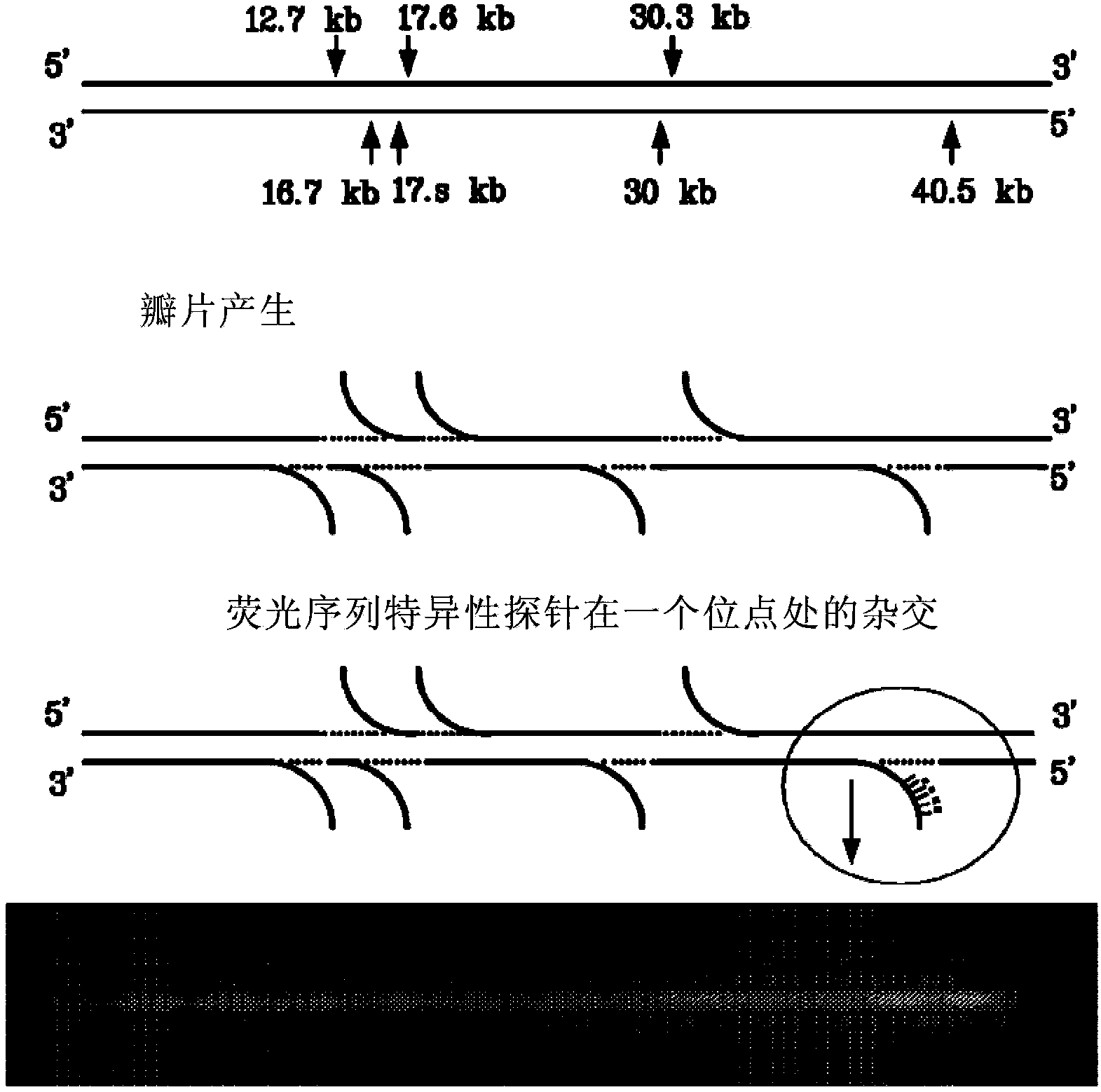
[0139] Example: Generation of Single-Stranded DNA Flaps on Double-Stranded DNA Molecules
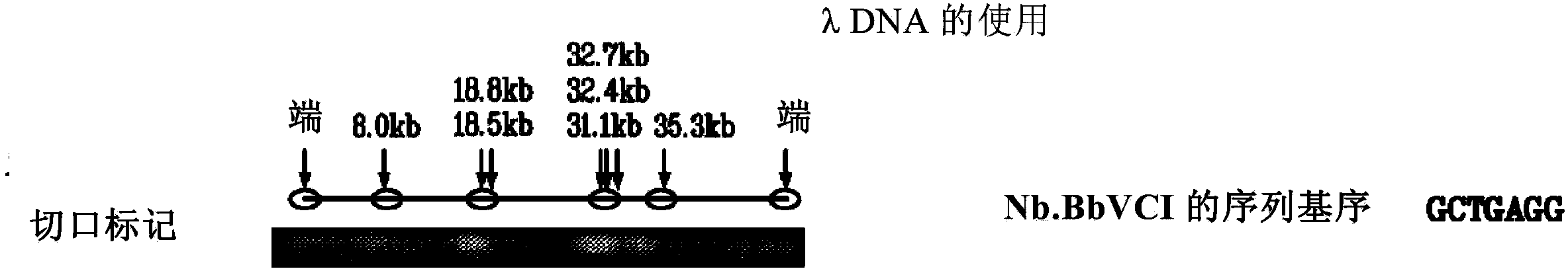
[0140] Genomic DNA samples were diluted to 50 ng for nicking reactions. Add 10 μL λDNA (50ng / μL) to a 0.2mL PCR centrifuge tube, then add 2 μL 10X NE Buffer #2 and 3 μL nicking endonucleases, including but not limited to Nb.BbvCI, Nb.BsmI, Nb.BsrDI, Nb. BtsI, Nt.AlwI, Nt.BbvCI, Nt.BspQI, Nt.BstNBI, Nt.CviPII. The mixture was incubated at 37°C for 1 hour.
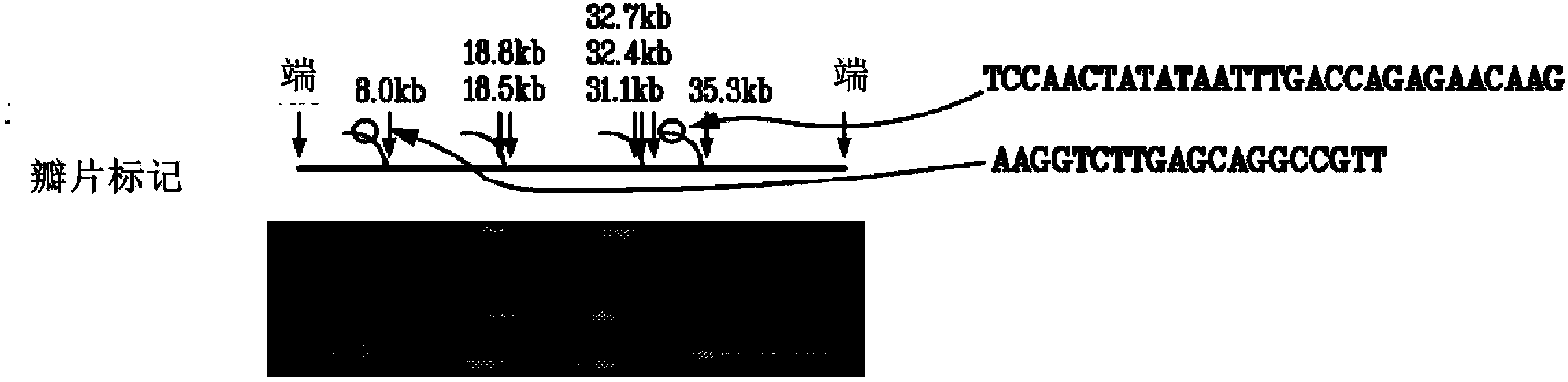
[0141] After the nicking reaction was complete, the experiment proceeded to limited polymerase extension at the nicking site to displace the 3' downstream strand and form a single-stranded flap. The flap generation reaction mix consisted of 15 μl of nicking product and 5 μl of incorporation mix containing 2 μl of 10X buffer, 0.5 μl of polymerase, and 1 μl of nucleotides at various concentrations from 1 μM to 1 mM. Enzymes include, but are not limited to, vent (exon-), Bst, and Phi29 polymerases. The flap generation reaction mixture...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap