Skeletal Muscle Chloride Channel Gene Mutant and Its Application
A chloride ion channel, skeletal muscle technology, applied in the fields of application, genetic engineering, plant gene improvement, etc., can solve the problems of congenital myotonia that need to be further deepened
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0050] Example 1 Target region capture sequencing to determine the mutation site
[0051] Sample collection: Collect peripheral blood from patient 1, patient 2 and their parents (peripheral blood from patient 1's parents cannot be collected), and use QIAamp DNA BloodMiNi Kit (Qiagen, Hilden, Germany) to extract genomic DNA as samples. Among them, patient 1 was diagnosed as congenital myotonia by the hospital, with typical myotonia, and his parents were normal; patient 2, a Cantonese, was diagnosed with tuberous sclerosis when he was 8 months old. He was 2 years and 8 months old (his parents had signed the informed consent form), and his clinical manifestations were: no hearing, no language ability, no walking, weak spine, and unable to stand upright for a long time. In addition, in this embodiment, the peripheral blood of 135 normal people was collected and their genomic DNA samples were extracted by the above-mentioned method, as a control.
[0052] The genomic DNA samples c...
Embodiment 2
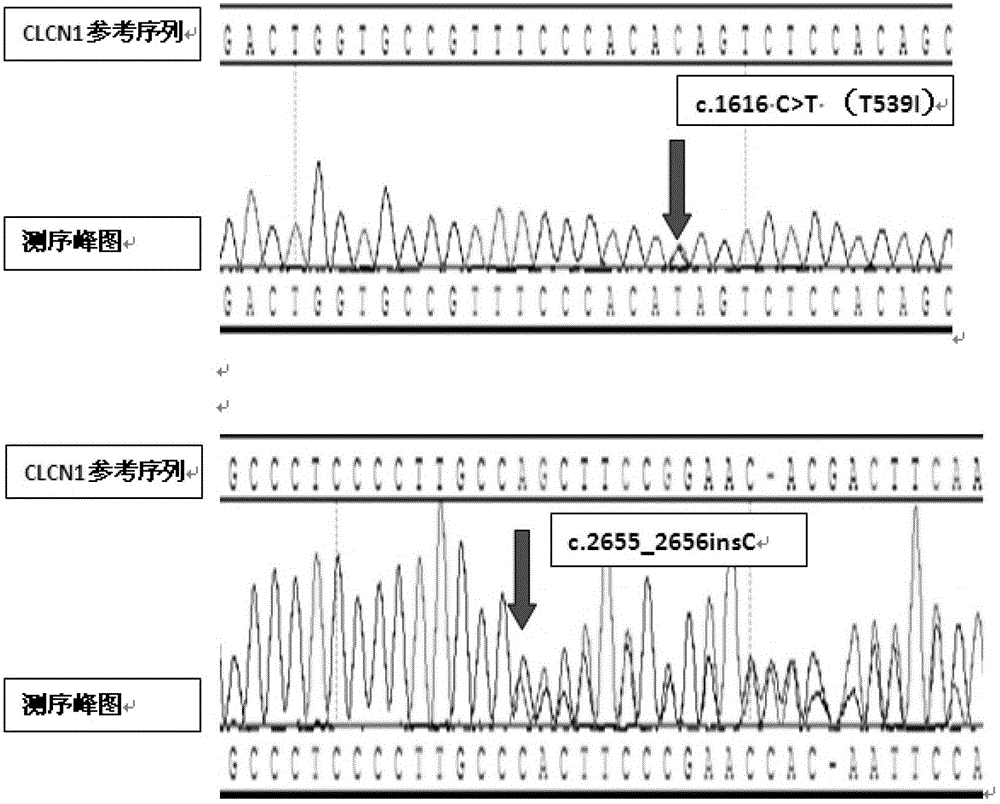
[0064] Example 2 Sanger method sequencing to verify the mutation sites of patient 1 and patient 2
[0065] Using the genomic DNA samples of patient 1 and 135 normal persons collected in Example 1, the CLCN1 gene mutation site of patient 1 obtained above was verified by Sanger sequencing according to the following steps:
[0066] 2.1 Primer design and PCR reaction
[0067] First, referring to the human genome sequence database hg19 / build36.3, the exon-specific primers of the skeletal muscle chloride ion channel gene (ie, CLCN1 gene) having the nucleotide sequence shown in SEQ ID NO: 3-32 were designed (see aforementioned Table 1). Wherein, the exon-specific primers are specific to 23 exons of CLCN1 and their adjacent intron sequences.
[0068] Then, configure the PCR reaction system of each genomic DNA sample according to the following ratio:
[0069]
[0070] Then, each PCR reaction system was carried out PCR reaction according to the following reaction conditions respec...
Embodiment 3
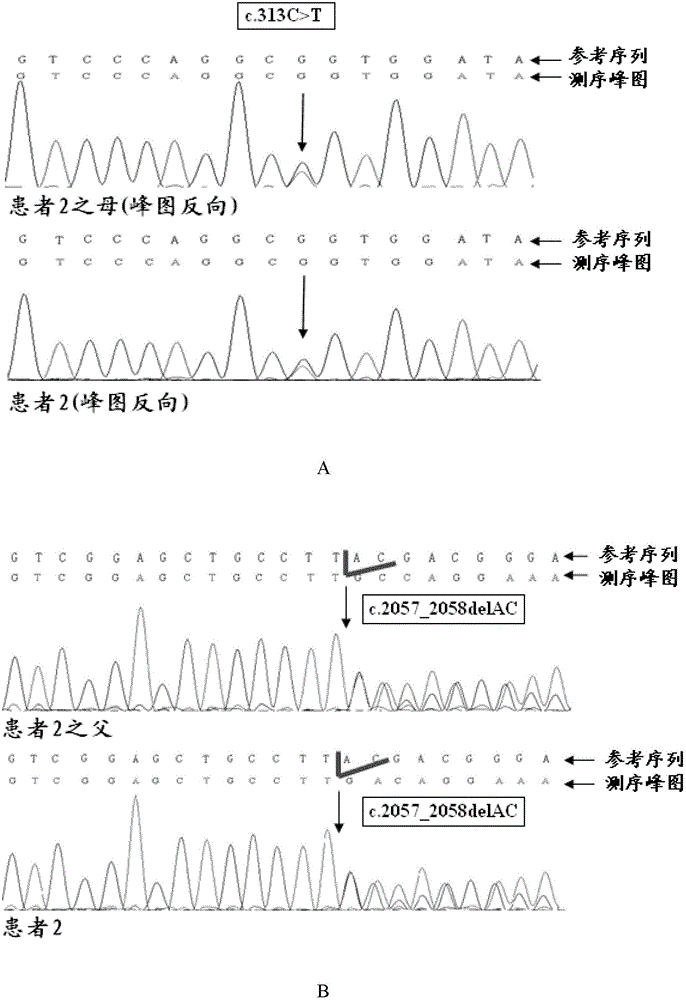
[0076] Example 3 Sanger method sequencing to verify the mutation sites of patients 1 and 2
[0077] Using the patient 2 and his parents collected in Example 1, and the genomic DNA samples of 135 normal people, according to the steps of Sanger method sequencing verification described in Example 2, the CLCN1 gene mutation site of the aforementioned patient 2 Sequencing verification by Sanger method. Wherein, the primers that PCR reaction adopts are different, and the PCR primers that the present embodiment adopts is:
[0078] (a) c.2057_2058delAC mutation site primer sequence
[0079] Upstream primer: GGCGCCTCTCCTGTTCCTT (SEQ ID NO: 33);
[0080] Downstream primer: GGAATTGCATGCAGGTCAAGG (SEQ ID NO: 34).
[0081] (b) Sequence of primers for p.Arg105Cys (i.e. c.313C>T) mutation site
[0082] Upstream primer: CGTTTCTTCCTAGATTGTATCCACC (SEQ ID NO: 35);
[0083] Downstream primer: GGAGGGGCGTGAGAAGTGG (SEQ ID NO: 36).
[0084] Thus, the alignment results of the skeletal muscle c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com