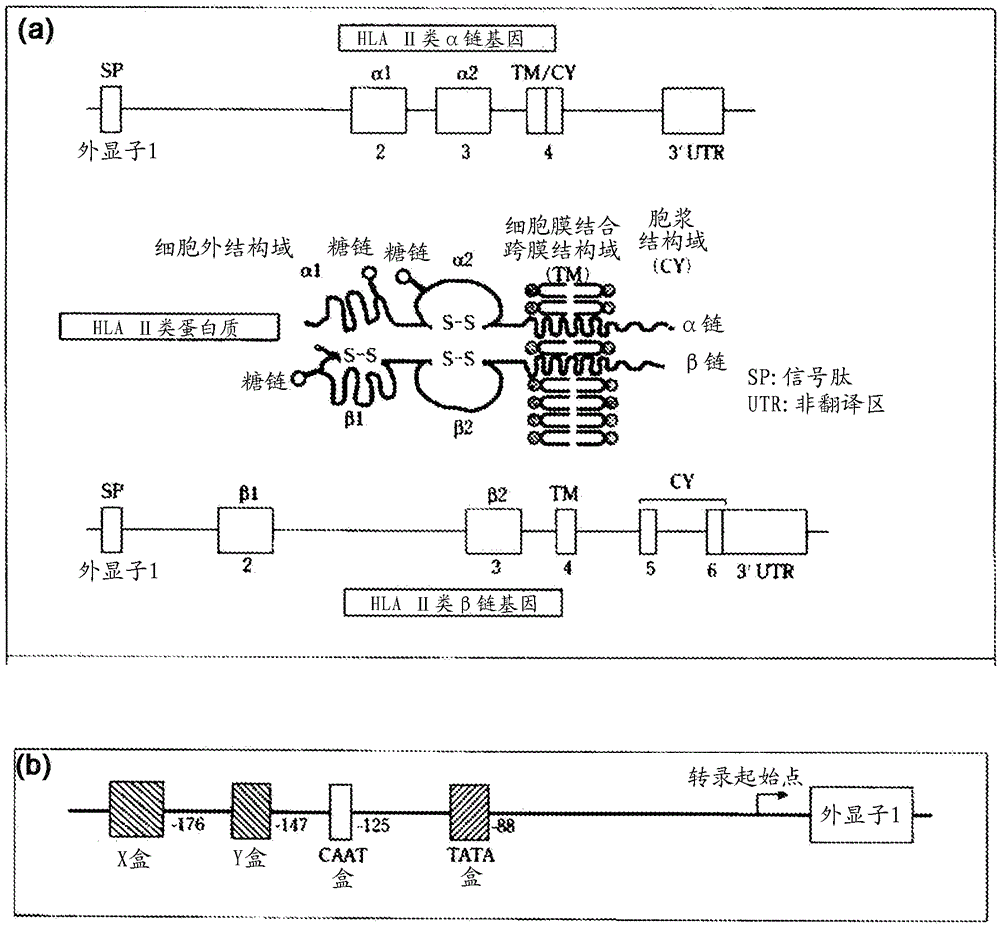
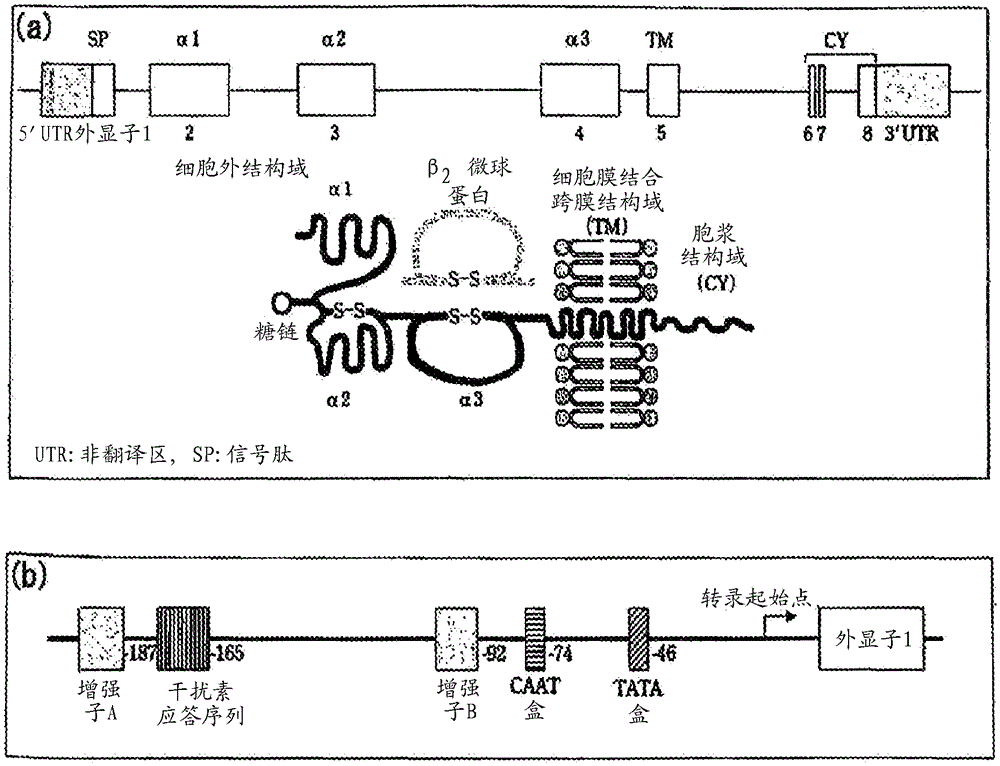
dna typing method and kit for hla gene
A technology of HLA-A and HLA-DRB1, which is applied in the field of DNA typing of HLA genes and kits, can solve problems such as difficult and high-precision HLA typing, inaccurate determination of cis-trans positional relationship, phase ambiguity, etc., and achieve high The effect of precision HLA matching
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1)
[0192] [experimental method]
[0193] 1. Using the extracted genomic DNA as a template, a PCR reaction was performed using specific primer sets for each HLA class I gene (refer to Table 1: sequence numbers 1 to 8). The specific sequence is as follows.
[0194] (1) Prime STAR GXL polymerase (TaKaRa) was used for PCR amplification. That is, in 50 ng of genomic DNA solution, add 4 μL of 5 × PrimeSTAR GXL buffer, 1.6 μL of dNTP solution, 4 μL (1 pmol / μL) of PCR primers, 0.8 μL of Prime STAR GXL polymerase, and inactivate Bacterial water to adjust the total amount of the reaction solution to 20 μL.
[0195] (2) After keeping warm at 94°C for 2 minutes, then repeat 30 times as a single process of 3 steps of 98°C for 10 seconds, 60°C for 20 seconds, and 68°C for 5 minutes. In addition, GeneAmp PCR System 9700 (Applied Biosystems) was used for this PCR amplification. After PCR, the amplification of PCR products was confirmed by agarose gel electrophoresis. Its electrophoresis is...
Embodiment 2)
[0205] [experimental method]
[0206] 1. Using the extracted genomic DNA as a template, PCR reactions were performed using specific primer sets for HLA class I and HLA class II genes (refer to Tables 1 to 4: sequence numbers 1 to 8, 9 to 22, and 31 to 50). The specific sequence is as follows.
[0207] (1) Prime STAR GXL polymerase (TaKaRa) was used for PCR amplification. That is, to 50 ng of genomic DNA solution, add 4 μL of 5 × PrimeSTAR GXL buffer, 1.6 μL of dNTP solution, 1 to 7 μL (4 pmol / μL) of PCR primers, 0.8 μL of Prime STAR GXL polymerase , and adjust the total amount of the reaction solution to 20 μL with sterilized water.
[0208] (2) After keeping warm at 94°C for 2 minutes, then repeat 2 steps of 98°C for 10 seconds and 70°C for 5 minutes as one process 30 times. In addition, GeneAmp PCR System 9700 (Applied Biosystems) was used for this PCR amplification. After PCR, the amplification of PCR products was confirmed by agarose gel electrophoresis. Its electrop...
Embodiment 3)
[0218] [experimental method]
[0219] 1. Genomic DNA was extracted using Buccal Cell DNA Extraction Kit, BuccalQuick (TRIMGEN).
[0220] 2. Genomic DNA extracted using Buccal Cell DNA Extraction Kit, BuccalQuick (TRIMGEN) was further purified by isopropanol and ethanol.
[0221] 3. Genomic DNA was extracted using QIAamp DNA Blood Mini Kit (QIAGEN).
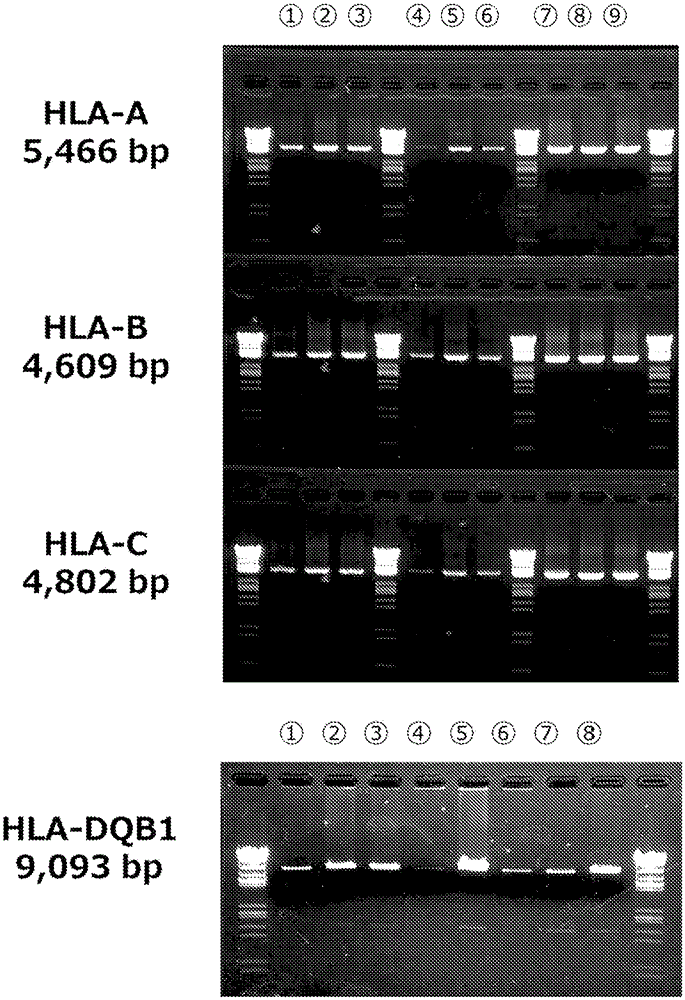
[0222] 4. Apply each of the 3 samples of genomic DNA extracted in items 1 to 3, and use the same experimental method as in Example 1 and Example 2 to apply HLA-A, HLA-B, HLA-C, and HLA-DQB1 specific primer sets (Refer to Table 1 and Table 4: Sequence Nos. 1 to 8, 29, 30, 48 to 50) PCR reactions were carried out. After PCR, the amplification of PCR products was confirmed by agarose gel electrophoresis. Its electrophoresis is shown in Figure 7.
[0223] [Experimental Results and Discussion]
[0224] Figure 7 Swimming lanes 1 to 3 show the amplification of PCR products under the condition of extraction with experimental ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com