Approaches to Altering Cell Fate
A cell and somatic cell technology, applied in the fields of changing cell chromatin structure, changing cell histone H3 methylation, changing cell heterochromatin protein HP1a expression and localization in cells, achieving good reproducibility and high efficiency , The method is simple and easy to implement
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Embodiment 1: Cloning of Gadd45 protein
[0055] Using the cDNA of the cell as a template, the primers shown in Table 1 were used to amplify the Gadd45a, Gadd45b and Gadd45g fragments respectively, and then recovered by agarose gel electrophoresis. Insert the PCR fragment after PmeI digestion at the multiple cloning site of the pMXs vector, connect, transform, pick bacteria, and sequence, and finally obtain the retroviral packaging plasmids of Gadd45a, Gadd45b and Gadd45g respectively.
[0056] Table 1 Primers for cloning Gadd45 protein fragments
[0057]
Embodiment 2
[0058] Embodiment 2: Gadd45 protein promotes reprogramming experiment
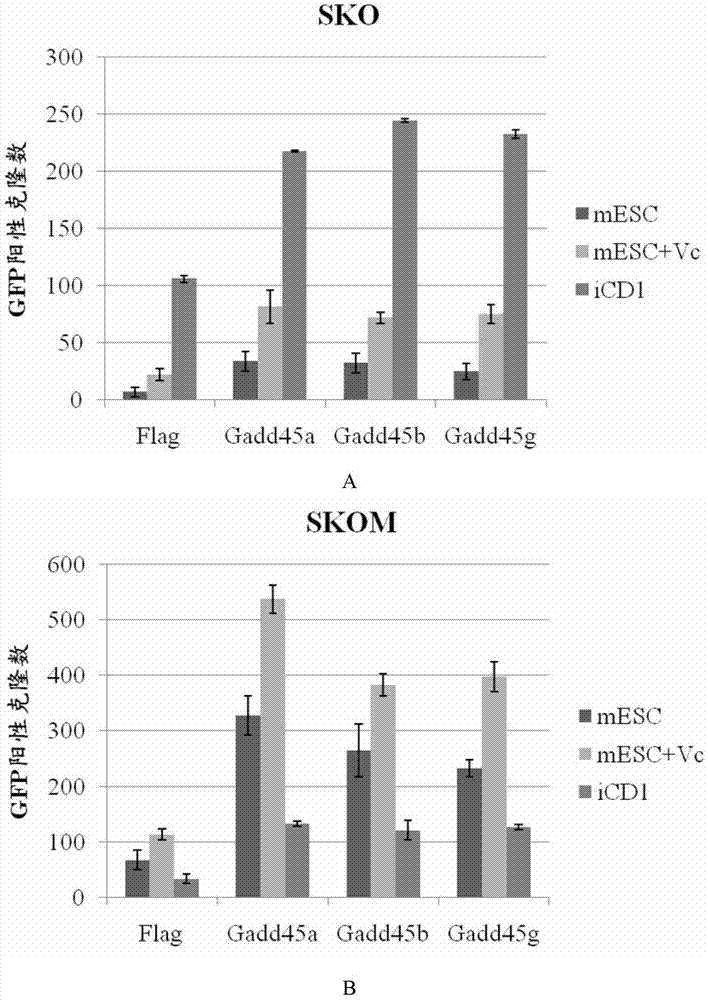
[0059] Retroviruses of reprogramming factors Sox2, Klf4, Oct4 and c-Myc as well as Gadd45a, Gadd45b and Gadd45g were packaged with Plat-E cell line. Collect the virus with a syringe, filter it into a centrifuge tube with a 0.45-micron filter, add an appropriate amount of fresh medium (about 12 ml in a 10 cm dish with a total volume), add polybrene and mix well, and it can be stored at 4 ° C or used immediately Infect. Another 24 hours later, the virus was collected in the same way as secondary infection.
[0060] After secondary infection, MEF cells are replaced with various induction media; commonly used is mESC medium or various compound molecules are added to mESC medium. After that, fresh medium needs to be replaced every day until the end of the experiment. If the efficiency of iPS is to be measured, OG2-MEF (GFP protein coupled to the endogenous Oct4 promoter) should be used. When iPS clones have...
Embodiment 3
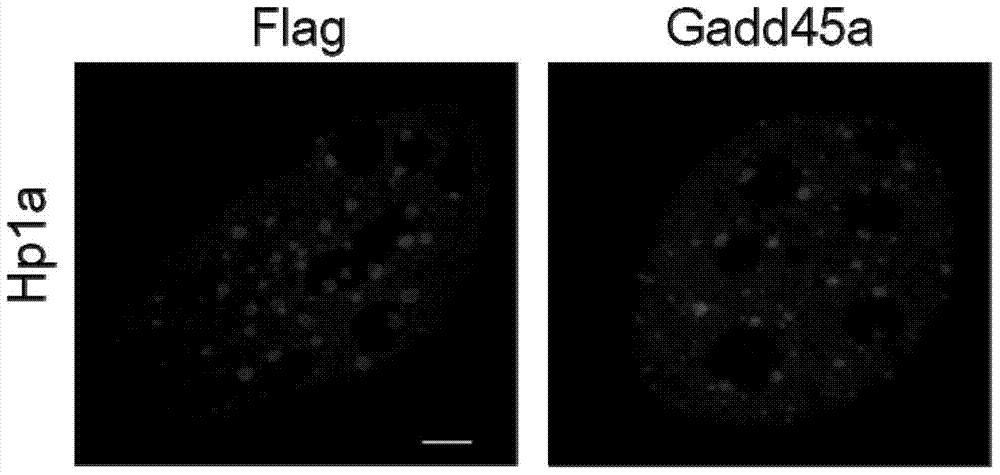
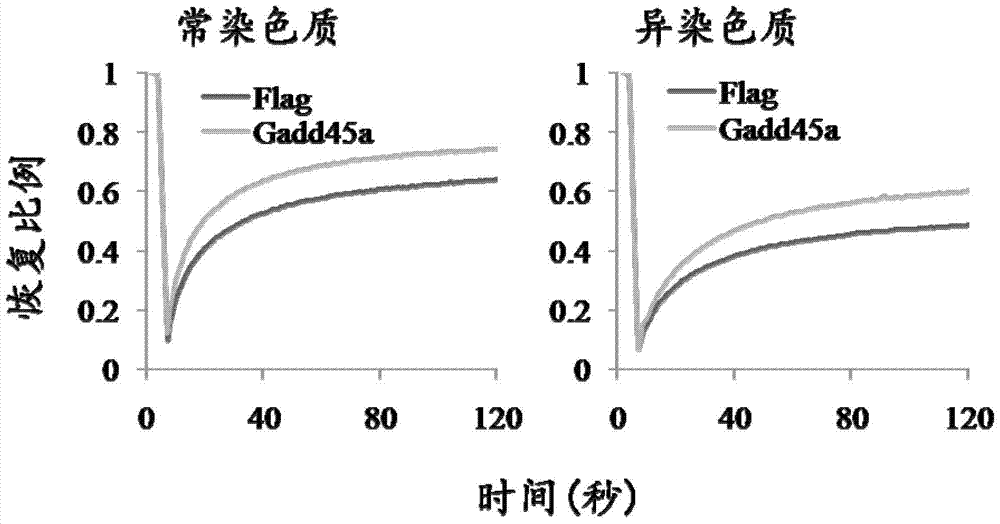
[0063] Example 3: Effect experiment of Gadd45a on heterochromatin protein HP1a
[0064] When analyzing the effect of Gadd45a on the heterochromatin protein HP1a, the retrovirus of Gadd45a must be packaged first to infect MEFs (any MEFs are acceptable). After culturing for 3 days, wash with PBS once, and fix with 4% paraformaldehyde at room temperature for half an hour. After fixation, absorb paraformaldehyde and replace with PBS. Wash once with glycine solution, then wash three times with PBS, and then permeabilize and block with a mixed solution of Triton X100 and BSA for 1 hour. After washing 3 times with PBS, incubate the primary anti-HP1a antibody for at least 1 hour; wash 5 times with washing solution and incubate the fluorescent secondary antibody. Finally, the nuclei were stained with DAPI, washed once with PBS, sealed on the glass slide with mounting medium, and then observed and photographed with a laser confocal microscope.
[0065] figure 2 The results of exper...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com