Detection of Streptococcus pneumoniae
A Streptococcus pneumoniae detection and labeling technology, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc., can solve the problem that there is no provision for Streptococcus specific detection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
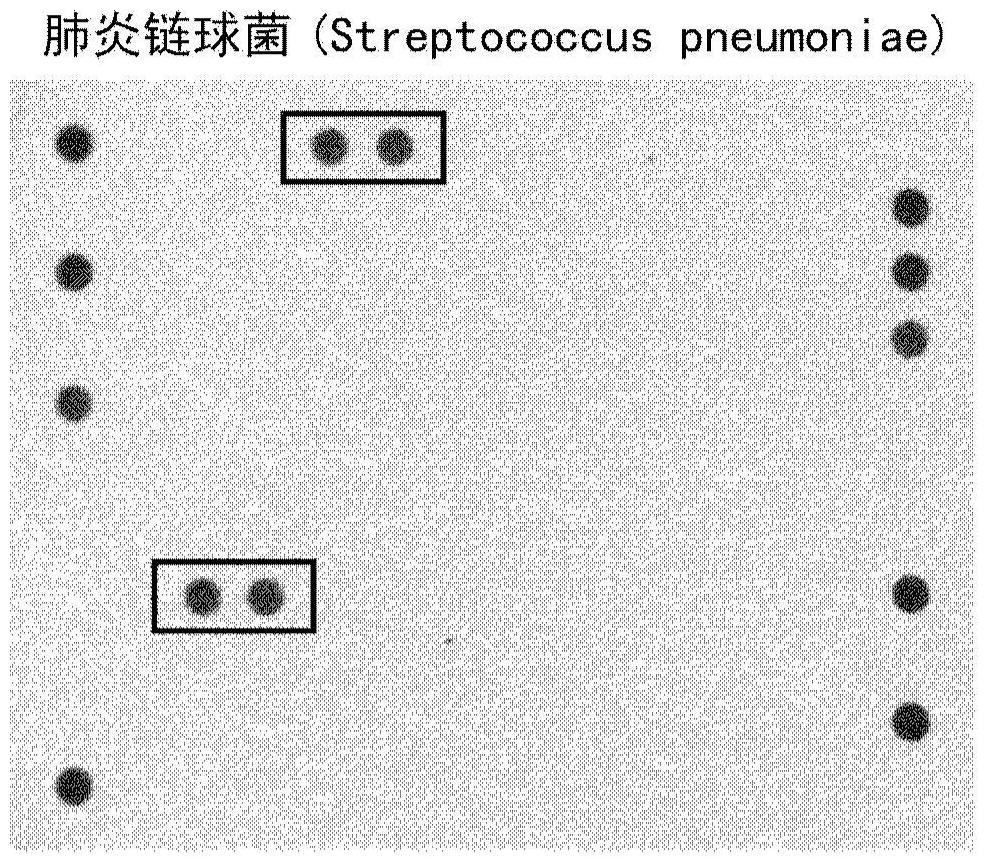
[0094] Samples of blood culture positive for S. pneumoniae were obtained and DNA was extracted therefrom by means of the NucliSENSeasy MAG kit from BioMerieux. The initial volume of blood culture used was 1 ml and the final volume after elution was 80 μl.
[0095] Next, take an elution volume of 5 μl and amplify it with a mixture of the following reagents:
[0096]
[0097]
[0098] Amplification products were denatured by incubation at 95°C for 10 minutes. Afterwards, in the presence of a standard hybridization solution, 5 μl of the denatured PCR product was mixed with the DNA containing the SEQ ID and SEQ ID Microarray incubation of probes.
[0099] After a series of standard washing and blocking steps, images were taken to obtain figure 1 A visualization of the final result is shown in .
Embodiment 2
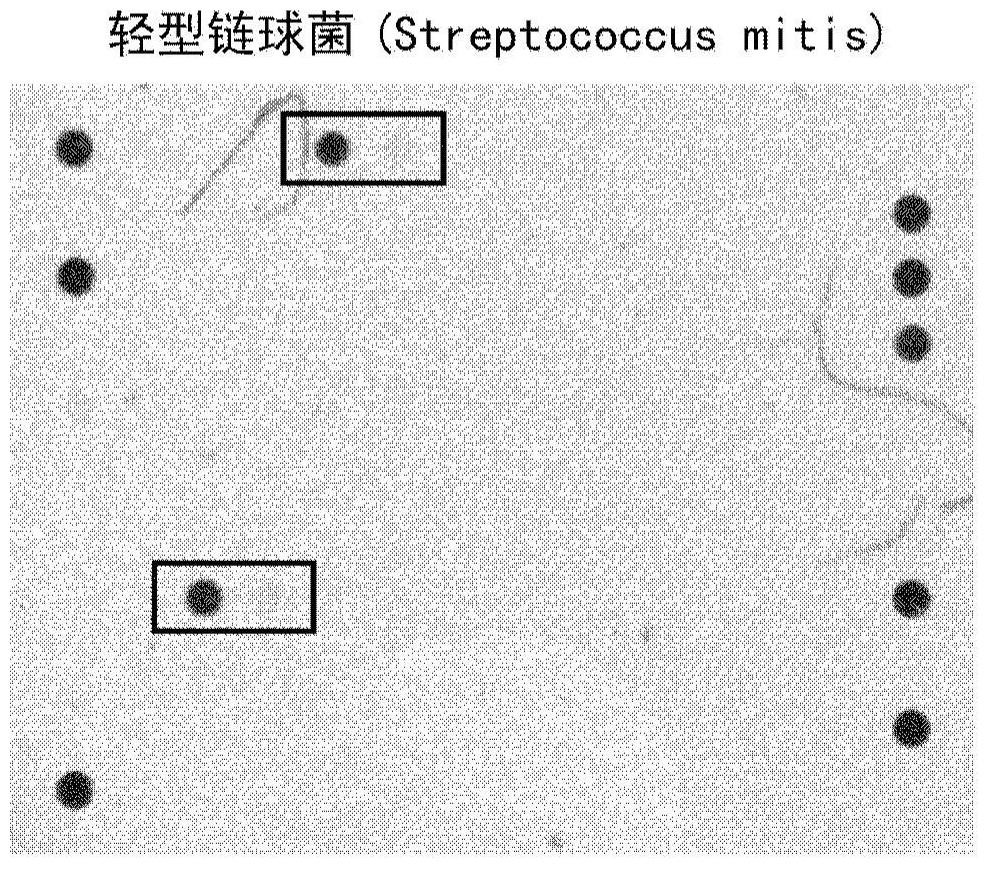
[0101] Samples of blood cultures positive for S. light were obtained and the same procedure described in Example 1 was performed for S. pneumoniae. exist figure 2 A visualization of the final result is shown in .
Embodiment 3
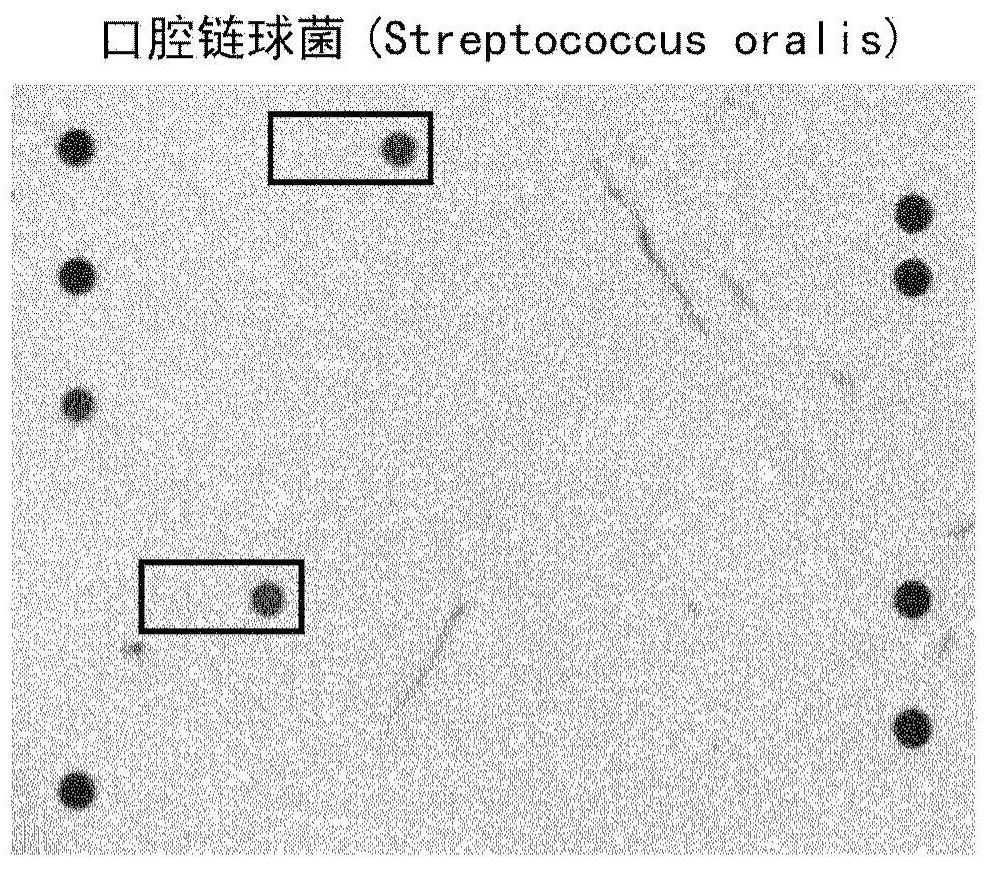
[0103] Samples of blood cultures positive for oral Streptococcus were obtained and the same procedure described in Example 1 was performed for Streptococcus pneumoniae.
[0104] exist image 3 A visualization of the final result is shown in .
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com