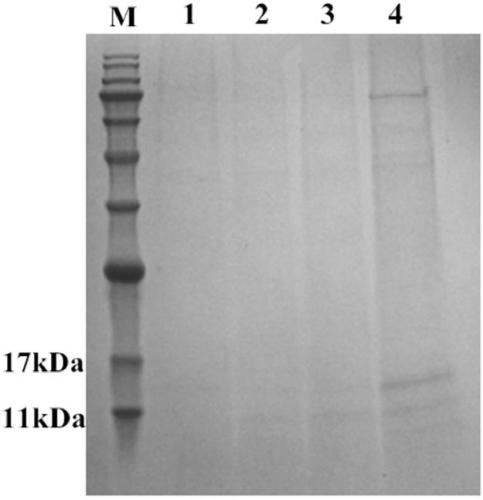
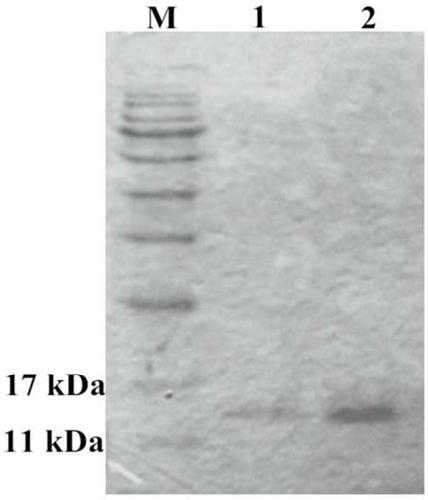
Antibacterial application of a gene hdiv-sarp19-i1 and its recombinant protein
A technology of hdiv-sarp19-i1, recombinant protein, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Hdiv-SARP19-I1 gene sequence and its optimization
[0056] The Hdiv-SARP19-I1 gene sequence was optimized according to the Hdiv-SARP19-I1 gene sequence (SEQ ID No.1) in the transcriptome of variegated abalone larvae and the codon preference of Pichia pastoris. In order to facilitate purification, a histidine purification tag (6*His tag) and an enterokinase tag were added to the N-terminus of the protein; in order to facilitate cloning, EcoR I and Not I restriction sites were introduced at the 5' and 3' ends, respectively . The optimized gene sequence is shown in SEQ ID No.2, which was synthesized by Shanghai Ruichu Biotechnology Co., Ltd. Among them, the restriction endonuclease EcoR I digestion site, histidine purification tag and enterokinase tag were introduced at bases 1-42, and Hdiv-SARP19 after removing the signal peptide and codon-optimized at 43-450 - In the coding region of I1 protein, 451-453 are stop codons, and 454-461 are restriction endonuclea...
Embodiment 2
[0057] Embodiment 2: Construction of expression vector
[0058] Pichia pastoris GS115 strain and expression vector pPIC9.0K were purchased from Shanghai Ruichu Biotechnology Co., Ltd. The synthesized target gene and vector pPIC9.0K were double-digested with restriction endonucleases EcoR I and Not I, and the digested products were subjected to agarose electrophoresis and the DNA fragments were recovered and ligated with a gel recovery kit. Specifically: add 5 μL of digested product of target gene, 7 μL of digested product of pPIC9.0K vector, 1 μL of T4 DNA ligase, 2 μL of 10×T4 DNA ligase buffer, add ddH2O to 20 μL in a sterile EP tube, mix well, and keep at 16°C Ligation for 12 hours; add 10 μL of the ligation product to 90 μL of competent Escherichia coli TOP10, mix well, heat shock at 42°C for 90 s for transformation, add 400 μL of LB liquid medium, shake and incubate at 37°C for 1 hour, and spread the bacterial solution on LB containing 25 μg / mL Zeocin Incubate overnight ...
Embodiment 3
[0059] Example 3: Transformation and strain screening
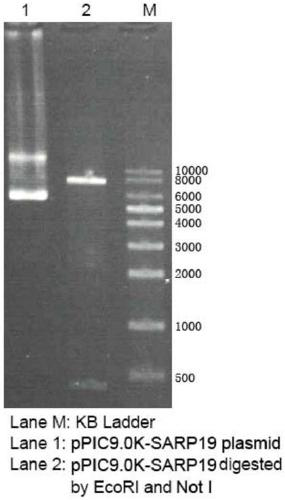
[0060]Linearize 5-10 μg of the recombinant plasmid pPIC9.0K-SARP19 using the restriction endonuclease KpnI, recover the linearized pPIC9.0K-SARP19 with a PCR product purification kit, add it to 90 μL of competent Pichia pastoris, mix well, After ice-bath for 5 minutes, transform by electric shock, add 1 mL of sorbitol and place at 30 °C for 2 h, spread the bacterial solution on YPD solid medium containing 100 μg / mL Zeocin, and incubate at 30 °C for 2 to 3 days. Select the colony on the transformation plate and culture it upside down on the YPD plate containing geneticin (G418) at a concentration of 0, 0.5 mg / mL, and 2 mg / mL at 30°C for 72 hours, and select the strain that grows well at 2 mg / mL as high copy strain. Streak the high-copy strains on MD and MM agar plates respectively, and observe after culturing at 30°C for 2 days. Mut+ transformants grow normally on both plates.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com