Hdiv-SARP19-I1 gene and antibacterial application of recombination protein of Hdiv-SARP19-I1 gene
A hdiv-sarp19-i1, recombinant protein technology, applied in the field of genetic engineering
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0055] Example 1: Hdiv-SARP19-I1 gene sequence and its optimization
[0056] The Hdiv-SARP19-I1 gene sequence was optimized according to the Hdiv-SARP19-I1 gene sequence (SEQ ID No.1) in the transcriptome of variegated abalone larvae and the codon preference of Pichia pastoris. In order to facilitate purification, a histidine purification tag (6*Histag) and an enterokinase tag were added to the N-terminus of the protein; in order to facilitate cloning, EcoRI and NotI restriction sites were introduced at the 5' and 3' ends, respectively. The optimized gene sequence is shown in SEQIDNo.2, which was synthesized by Shanghai Ruichu Biotechnology Co., Ltd. Among them, the restriction endonuclease EcoRI cutting site, histidine purification tag and enterokinase tag were introduced at bases 1 to 42, and Hdiv-SARP19- In the coding region of the I1 protein, 451-453 are stop codons, and 454-461 are restriction endonuclease NotI cleavage sites.
Embodiment 2
[0057] Embodiment 2: Construction of expression vector
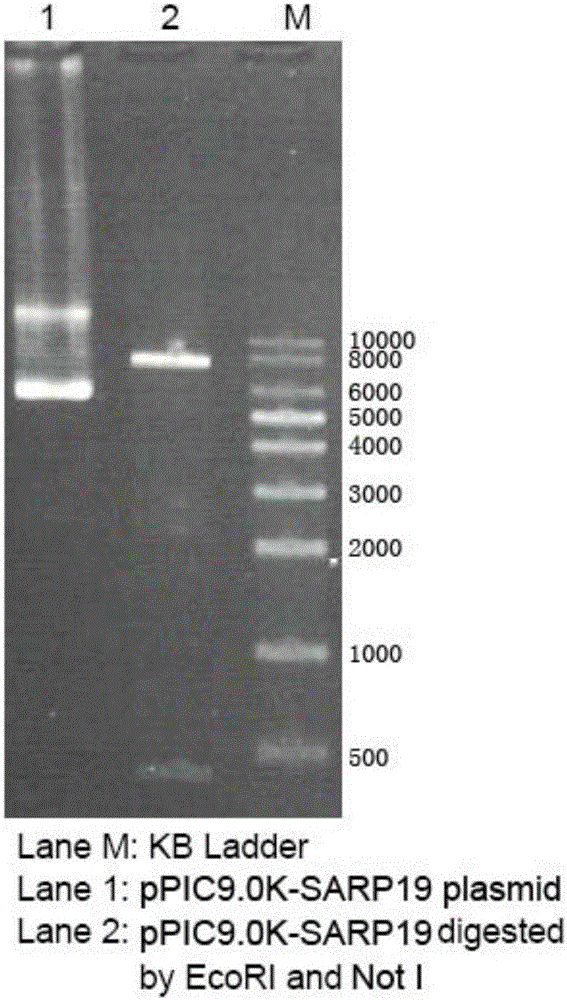
[0058] Pichia pastoris GS115 strain and expression vector pPIC9.0K were purchased from Shanghai Ruichu Biotechnology Co., Ltd. The synthesized target gene and vector pPIC9.0K were double-digested with restriction endonucleases EcoRI and NotI, and the digested products were subjected to agarose electrophoresis and the DNA fragments were recovered and ligated with a gel recovery kit. Specifically: add 5 μL of digested product of the target gene, 7 μL of digested product of pPIC9.0K vector, 1 μL of T4 DNA ligase, 2 μL of 10×T4 DNA ligase buffer, add ddH2O to 20 μL in a sterile EP tube, mix well, and keep at 16°C Ligation for 12 hours; add 10 μL of the ligation product to 90 μL of competent Escherichia coli TOP10, mix well, heat shock at 42°C for 90 s for transformation, add 400 μL of LB liquid medium, shake and incubate at 37°C for 1 hour, and spread the bacterial liquid on LB solids containing 25 μg / mL Zeocin Incubate ove...
Embodiment 3
[0059] Example 3: Transformation and strain screening
[0060] Linearize 5-10 μg of the recombinant plasmid pPIC9.0K-SARP19 using the restriction endonuclease KpnI, recover the linearized pPIC9.0K-SARP19 with a PCR product purification kit, add it to 90 μL of competent Pichia pastoris, mix well, After ice-bath for 5 minutes, electroshock transformation, add 1 mL of sorbitol, place at 30°C for 2 hours, spread the bacterial solution on YPD solid medium containing 100 μg / mL Zeocin, and incubate at 30°C for 2 to 3 days. Select the colony on the transformation plate and culture it upside down on the YPD plate containing geneticin (G418) at a concentration of 0, 0.5 mg / mL, and 2 mg / mL at 30°C for 72 hours, and select the strain that grows well at 2 mg / mL as high copy strain. Streak the high-copy strains on MD and MM agar plates respectively, and observe after culturing at 30°C for 2 days. Mut+ transformants grow normally on both plates.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com