A Bayesian Approach for Joint Estimation of Genomic Breeding Values for Continuous Traits and Threshold Traits
A technology of joint estimation and breeding value, applied in the Bayesian field, can solve the problem of lack of joint analysis, and achieve the effect of increasing the prediction accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
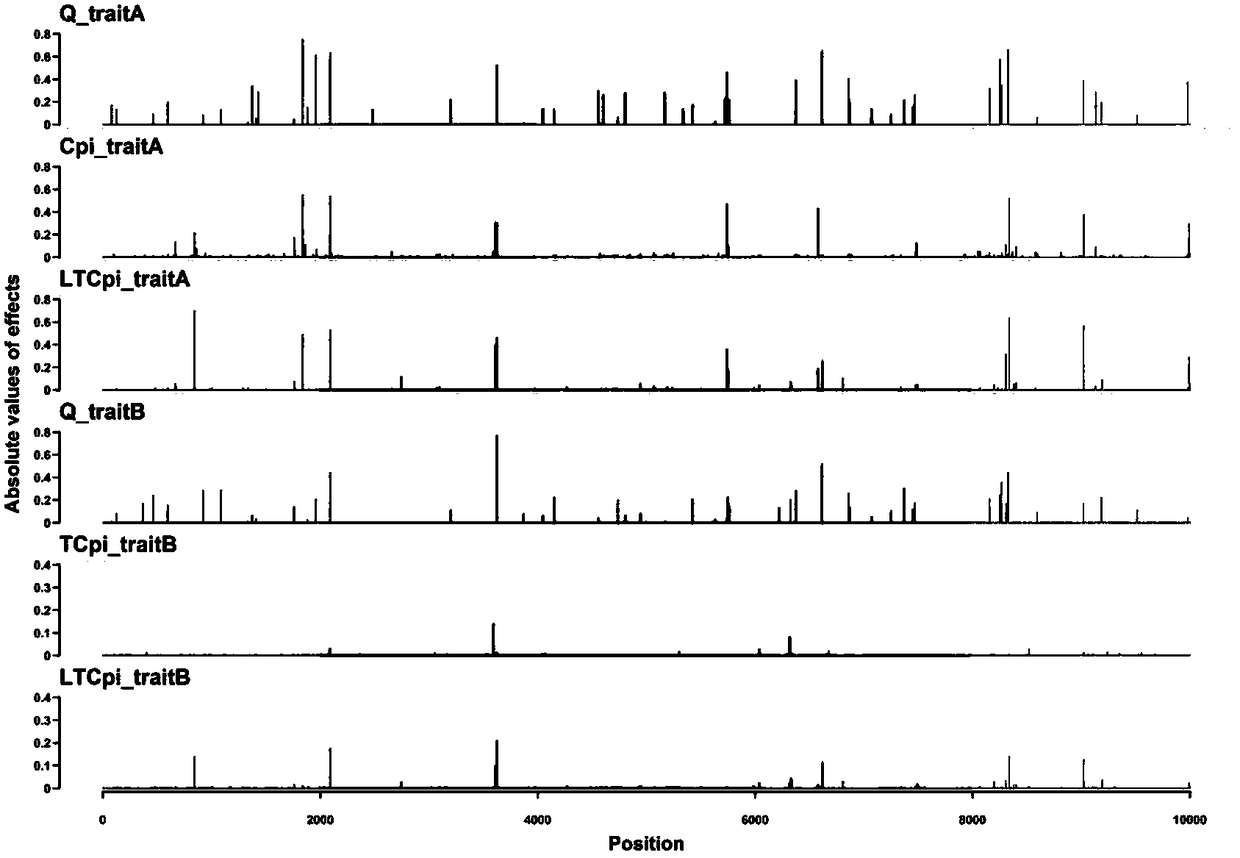
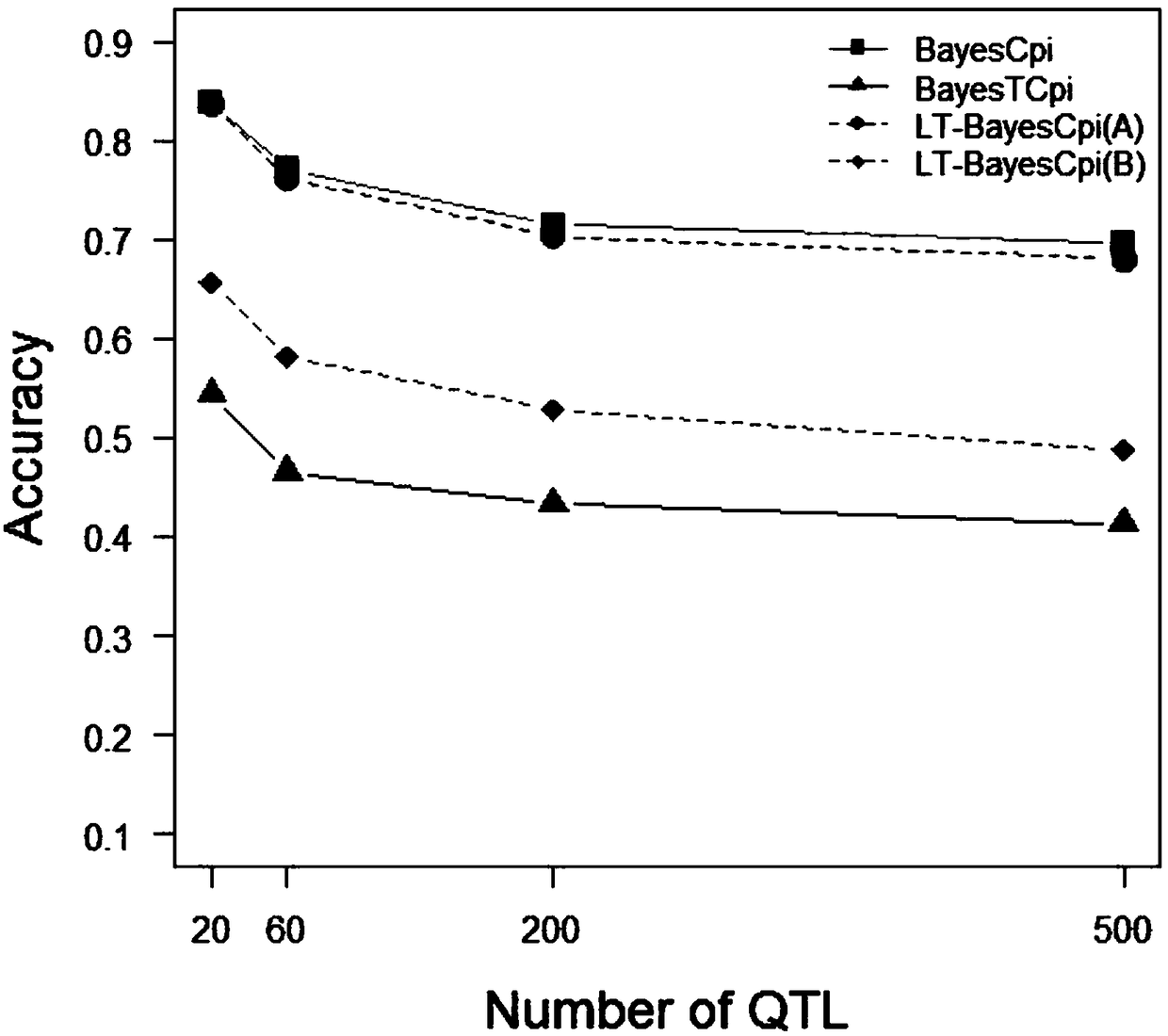
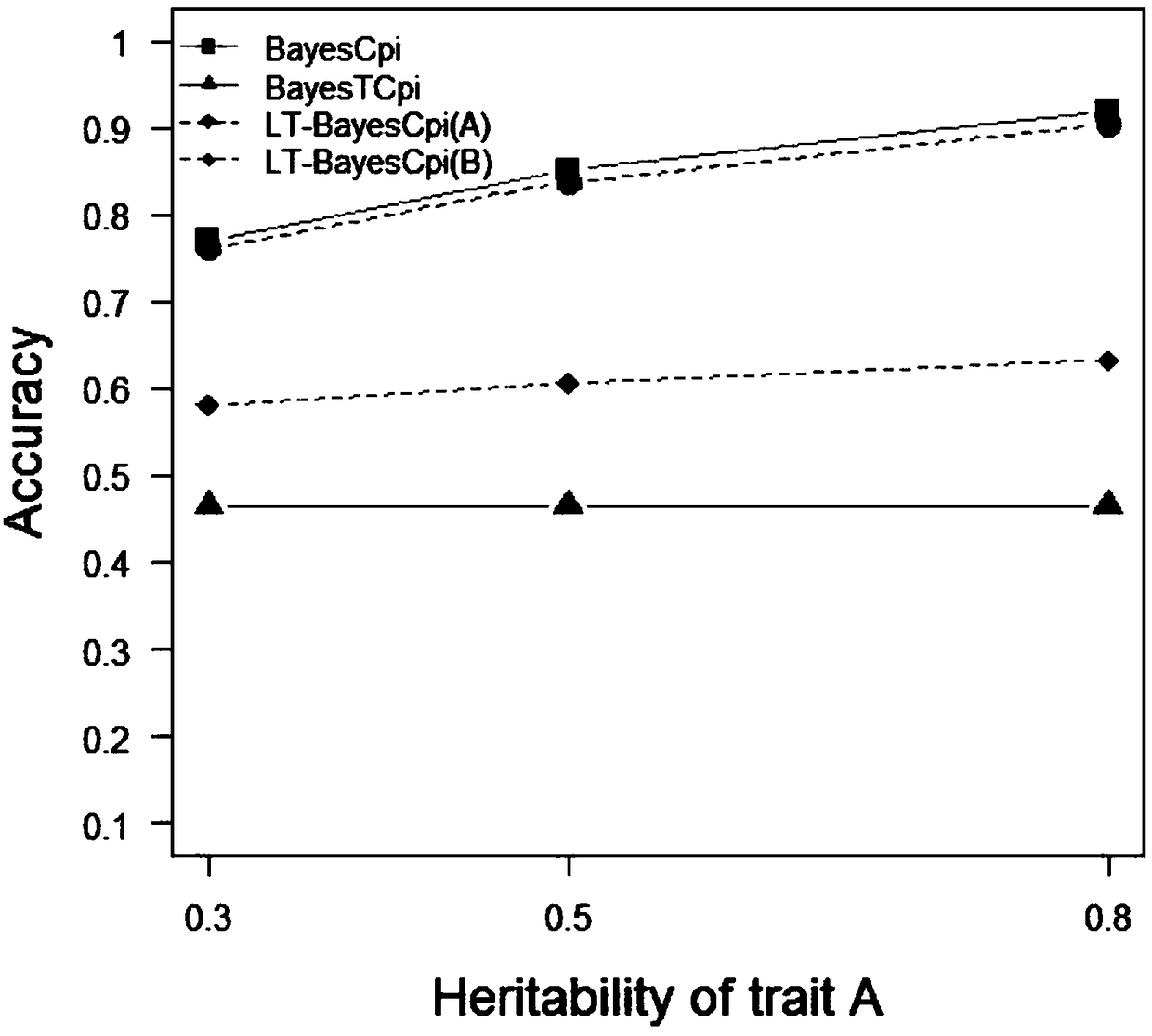
[0061] This example proposes a new Bayesian method based on the line-threshold model, called LT-BayesCπ, for joint analysis of continuous traits and threshold traits.
[0062] 1. Method
[0063] 1.1 Model
[0064] let y′ 1 ={y 1,i}, i=1, 2,..., n is the observation value vector of continuous traits, y' 2 ={y 2,i}, i=1, 2,..., n is the threshold trait observation value vector, l'={l i}, i=1, 2, ..., n is the latent variable vector associated with the threshold trait. The line-threshold model is:
[0065] where β 1 , β 2 is the fixed effect vector; g 1 , g 2 is the SNP effect vector; e 1 、e 2 is the random residual vector; x 1 、x 2 for β 1 , β 2 The association matrix of ; Z is the genotype indicator matrix, where assignments 0, 1, and 2 correspond to 11, 12, and 22 of the genotype, respectively. Let v'=[y' 1 , l′], given β and g, v obeys the following distribution:
[0066]
[0067] where β'=[β' 1 ,β′ 2 ], g'=[g' 1 , g′ 2 ],
[0068] Then given β,...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com