Caffe based extraction method of stack denoising self-encoding gene information characteristics
A gene feature and feature extraction technology, applied in the field of bioinformatics
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
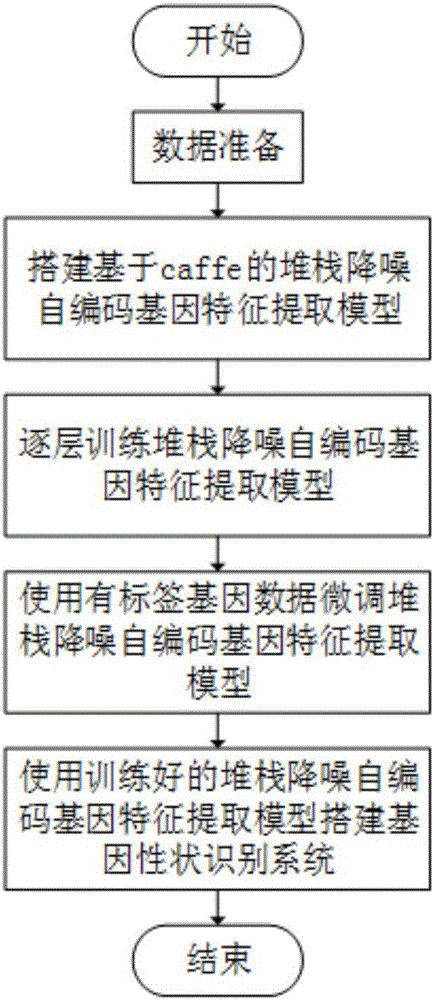
[0023] Data preparation in step S1: first prepare the gene information data used for training the model, where the gene information data uses genes with the same name from different individuals, assuming that there are n gene sample data from different individuals. Visualize the base sequence of the gene of the n people, and the visualization result is genetic data in n image formats;
[0024] Suppose the image pixel size is p 0 ×q 0 , and then set the n image pixels to a fixed size p×q. Use the convert_imageset tool provided by Caffe to convert the n image samples into a database file suitable for Caffe. The database file format is leveldb or lmdb, preferably lmdb.
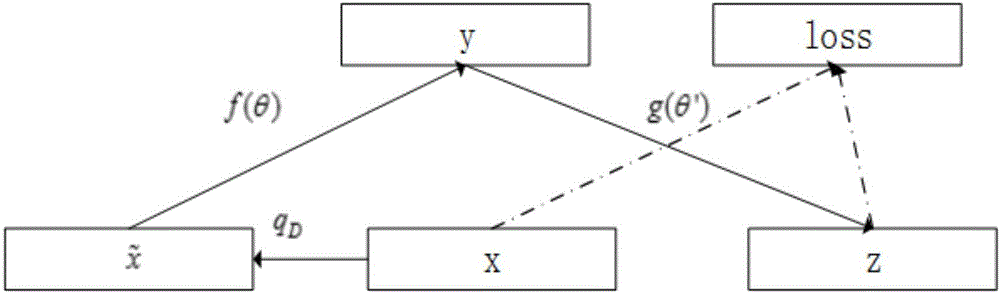
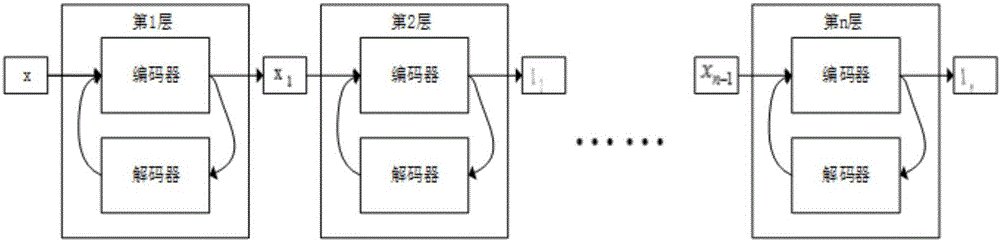
[0025] Step S2 is to build a stack noise reduction self-encoding gene feature extraction model based on Caffe. The basic unit of the model is a noise-reduction self-encoding model, and a gene feature extraction model is composed of several basic units of the noise-reduction self-encoding model stacked together...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com