A circRNA PSY1-circ1 involved in lycopene biosynthesis
A tomato and plant technology, applied in the field of nucleic acid, can solve the problem of unclear regulation mechanism of synthetic gene expression, and achieve the effect of being beneficial to genetic regulation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0058] Example 1. Bioinformatics and molecular biology identification of circRNA PSY1-circ1
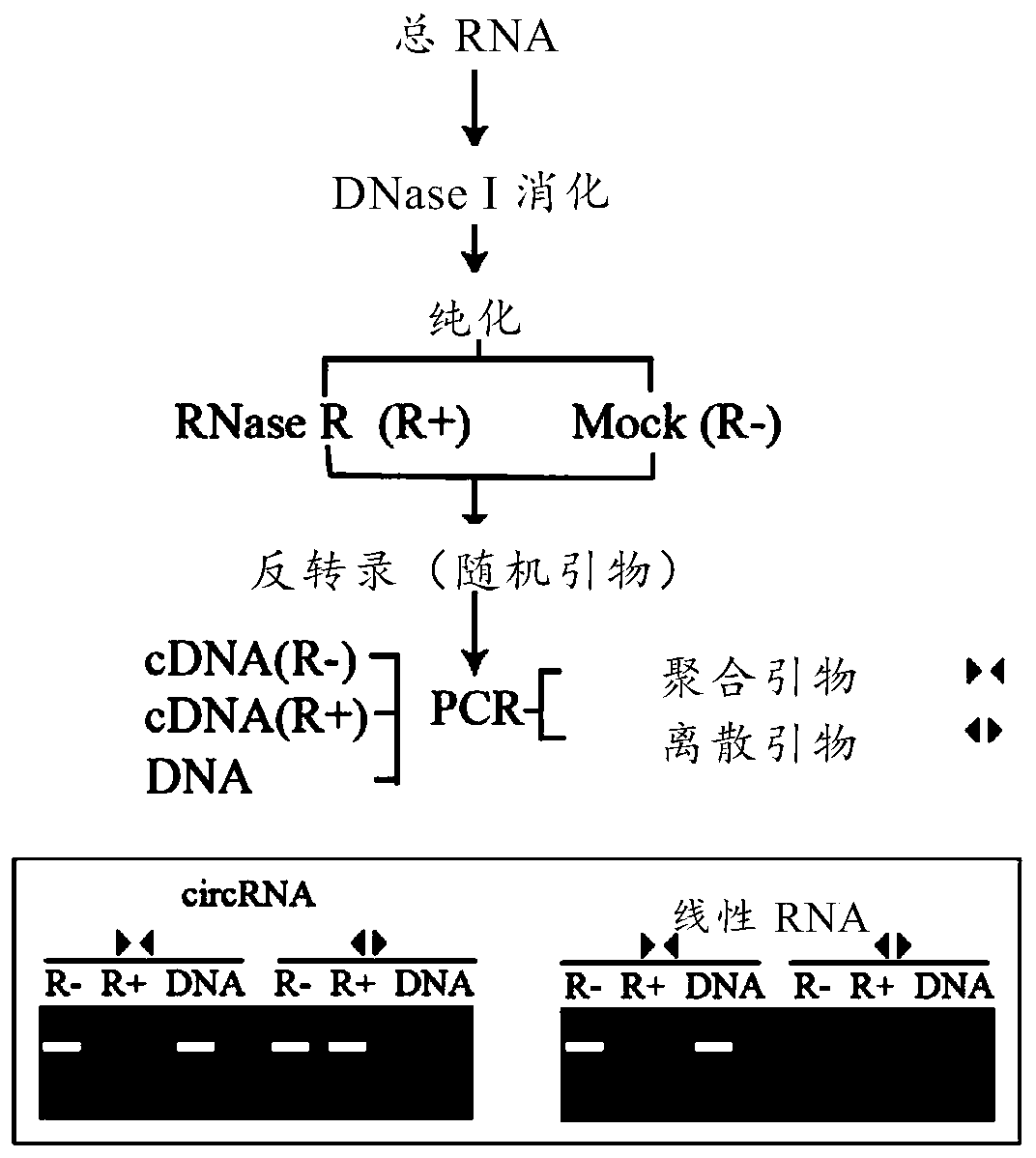
[0059] Deep sequencing of circular RNAs in tomato fruit : Trizol (Life technologies) was used to extract the total RNA of wild-type tomato (Solanum lycopersicum) variety (Ailsa Craig, hereinafter referred to as AC) in the green ripening (MG), breaking color (BR) and red ripening (BR+6) fruits , and then digested with DNase I (Fermentas) to remove residual genomic DNA. Afterwards, Ribo-Zero Magnetic Kit-plant leaf (Epicentre) was used to remove ribosomal RNA, and then RNase R (Epicentre) was used to treat at 37°C for 1 hour to digest linear RNA. The circRNA library was built with Illumina's RNA LT Sample Prep Kit v2, the insert length is 120-250bp; HiSeq2500 sequencer (2*100bp) for bidirectional sequencing, using 100bp paired-end reads (Lingneng Biotechnology (Shanghai) Co., Ltd.), valid for each sample The amount of data is more than 4Gbase.
[0060] Extraction and analysis of...
Embodiment 2
[0065] Example 2. Analysis of expression patterns of circRNA PSY1-circ1 and its source genes in tomato fruits at different maturity stages
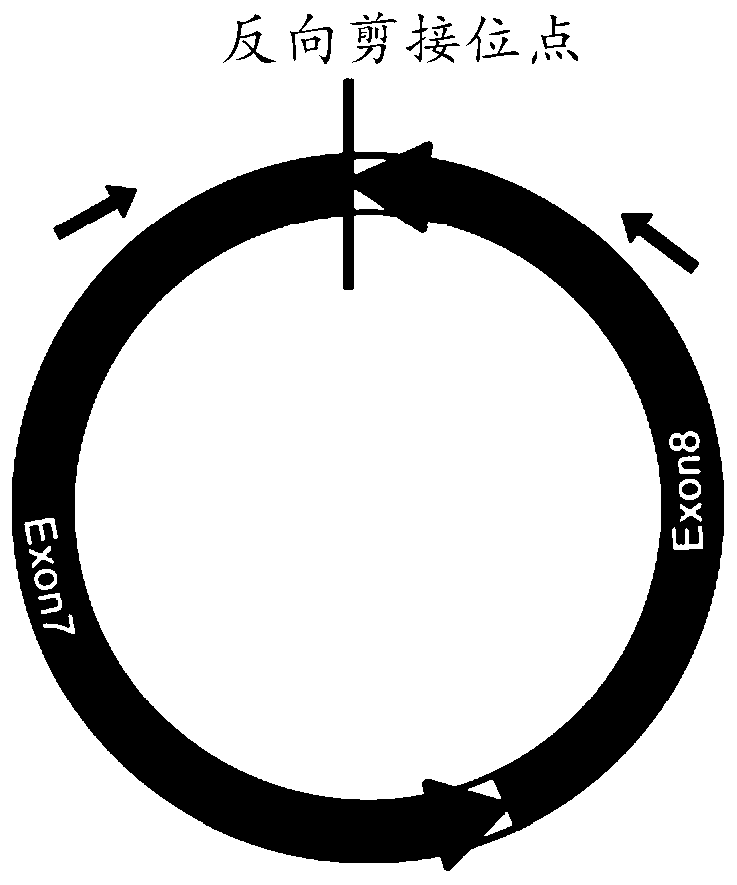
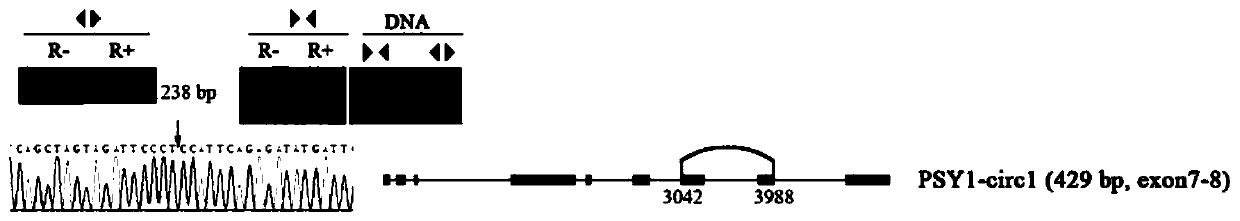
[0066] Design of special primers for identification of circRNA (real-time PCR method) : Due to the existence of selective circularization, the same gene can produce multiple circRNAs with overlapping regions, so the design of discrete primers (forward primers or reverse primers) to identify circRNAs spans the reverse splicing site (primers span the site The length of the 3' end of the point is 3-6bp) to ensure the specificity of the amplification, see the schematic diagram for details ( Figure 4 ). The rest of the design concept is the same as that of ordinary real-time PCR.
[0067] The primers obtained from this design concept for real-time PCR amplification of circRNA PSY1-circ1 are discrete primers, in which the forward primer is TCAGCTAGTAGATTCCCTCCATTC (SEQ ID NO: 8), and the reverse primer is CCTTTGATTCAGGGGCGATAC (SEQ ID NO: 9...
Embodiment 3
[0071] Example 3. Construction and genetic transformation of circRNA PSY1-circ1 overexpression vector
[0072] Construction of circRNA overexpression vector : Using the genomic DNA of wild-type tomato (AC) as a template, amplify the fragment (fragment A) containing the exon and flanking sequence (upstream 281bp and downstream 681bp) to be circularized (fragment A), and amplify part of the downstream sequence (517bp, fragment B ). Then Fragment A and Fragment B were connected in the opposite direction and constructed into the expression cassette of the pCAMBIA1301 vector ( Figure 6 ). Fragment A and fragment B in the carrier form a secondary structure due to partial sequence complementarity, which is the key to promoting the production of circRNA in large quantities. In addition, the vector retains the splicing sites and some flanking sequences on both sides of the exon to be circularized, ensuring precise splicing.
[0073] genetic transformation : The vector prepared ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com