A method for quantitative detection of dna point mutations
A quantitative detection method and detection method technology, applied in the direction of biochemical equipment and methods, microbial measurement/inspection, etc., can solve the problems of high detection cost, incomplete clamping of PNA-LNA clamped PCR, easy pollution of two-step nested PCR, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
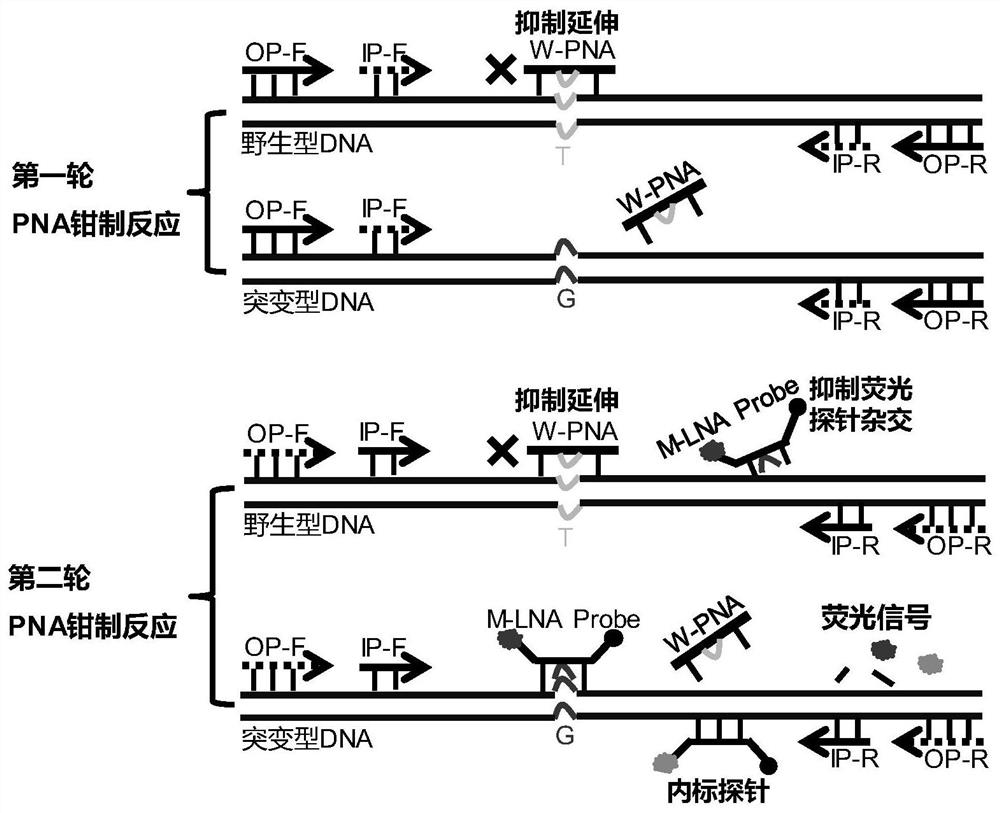
[0056] The present invention establishes a highly sensitive point mutation detection method, in order to evaluate the detection of clinical samples by the method. We applied this method to the detection of EGFR L858R mutation sites related to targeted drugs in plasma and tissue samples of non-small cell lung cancer.
[0057]EFGR is a common driver gene in non-small cell lung cancer, and its mutations mainly occur in exons 18 / 19 / 20 / 21. EGFR L858R (NCBI accession number: NG_007726.1) is the 858th amino acid encoded by exon 21 of the EGFR gene, which is changed from leucine to arginine. This mutation can cause the activation of the downstream pathway of EGFR, and then lead to the occurrence of tumors . Iressa (Iressa) and Tarceva (Tarceva) are common EGFR tyrosine kinase inhibitors, and are currently commonly used targeted drugs for the treatment of non-small cell lung cancer. A large number of studies have shown that EGFR L858R is one of the sensitive mutations of Iressa and T...
Embodiment 2
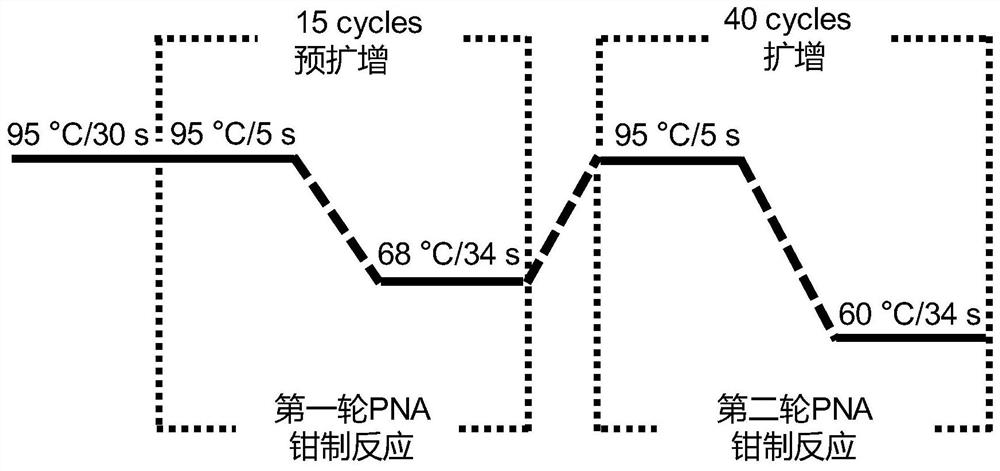
[0066] The method of the present invention is the development and improvement of the PNA-LNA clamped PCR technique, so the method of the present invention and the PNA-LNA clamped PCR technique have been compared with each other. The difference between the two methods in terms of amplification sensitivity and mutation detection sensitivity was mainly compared.
[0067] 1. The primers and sequences of the experimental group and the matched group 1 of the present invention are the same as in Example 1;
[0068] 2. Preparation of standard substances with known content: NCI-H1975 cells containing mutant EGFR L858R and H293T cells containing wild-type EGFR L858R were purchased from American Type Culture Collection (Rockville, USA);
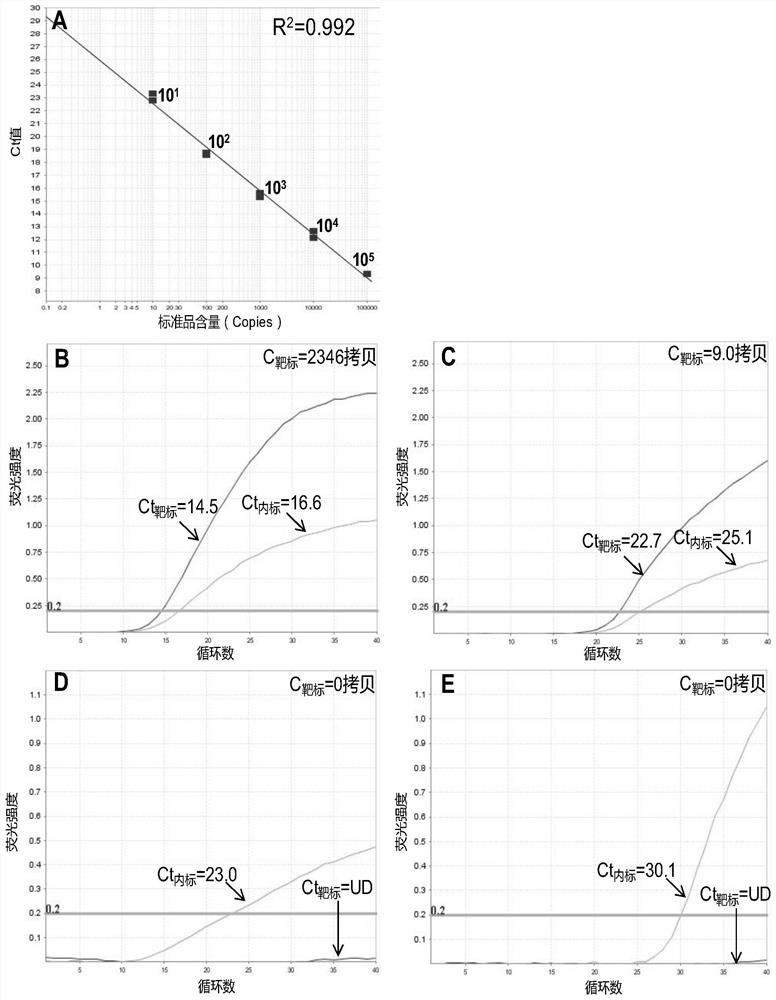
[0069] The EGFR L858R wild-type and mutant DNA in the cells were quantified by digital PCR, and used as standards of known content of the wild-type and mutant DNA, respectively. The EGFR L858R mutant cell DNA of known concentration was diluted to 10 by...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com