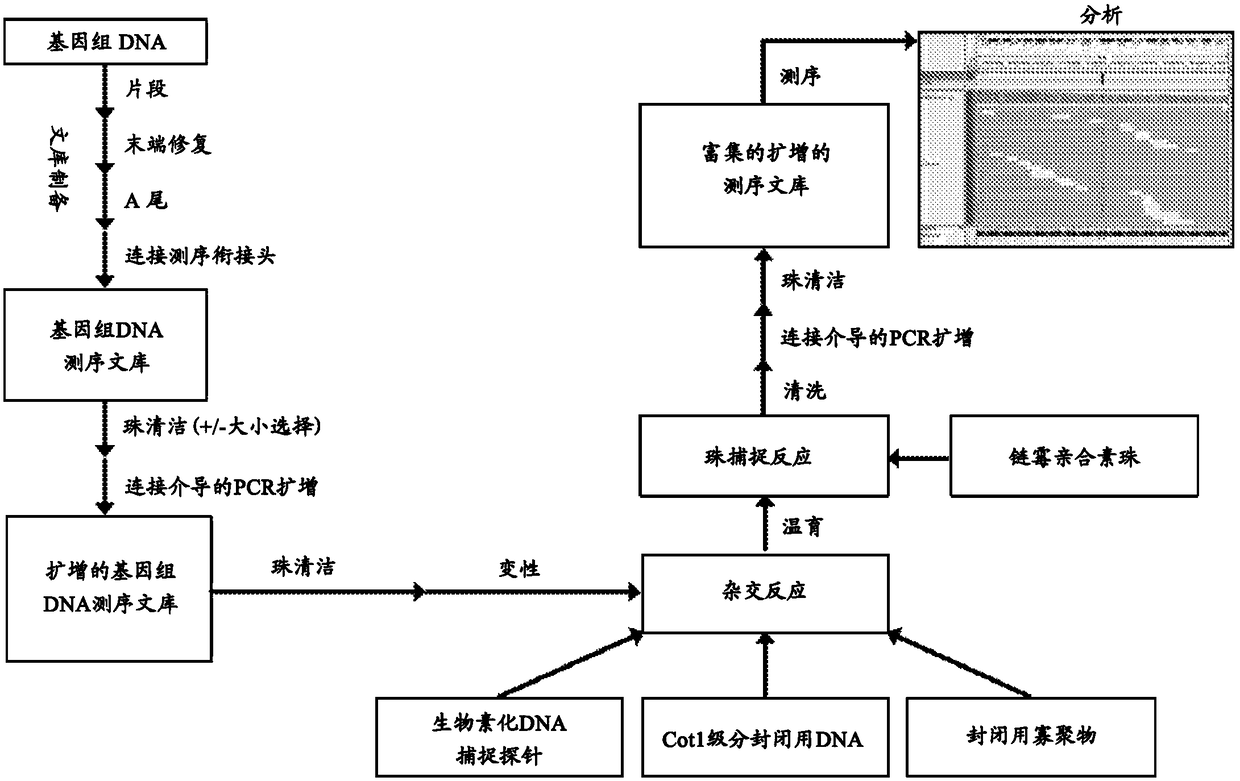
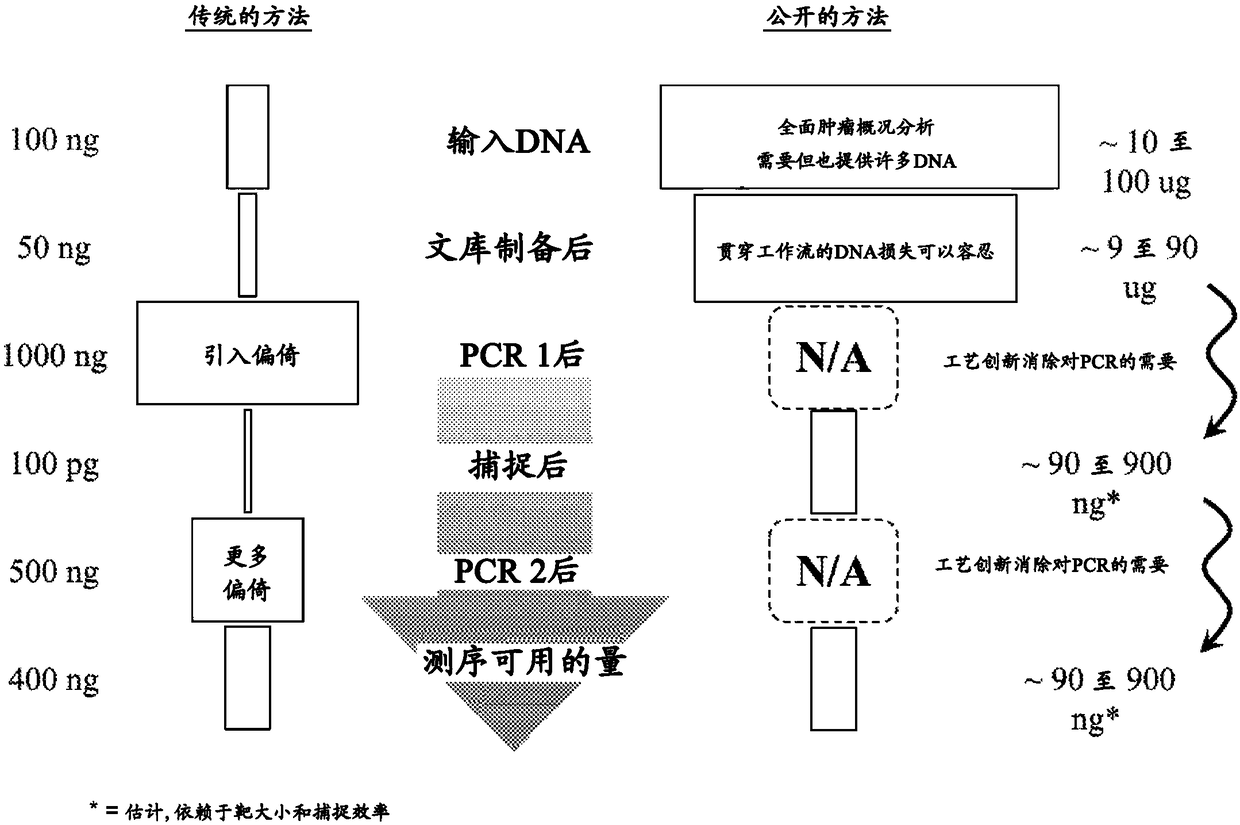
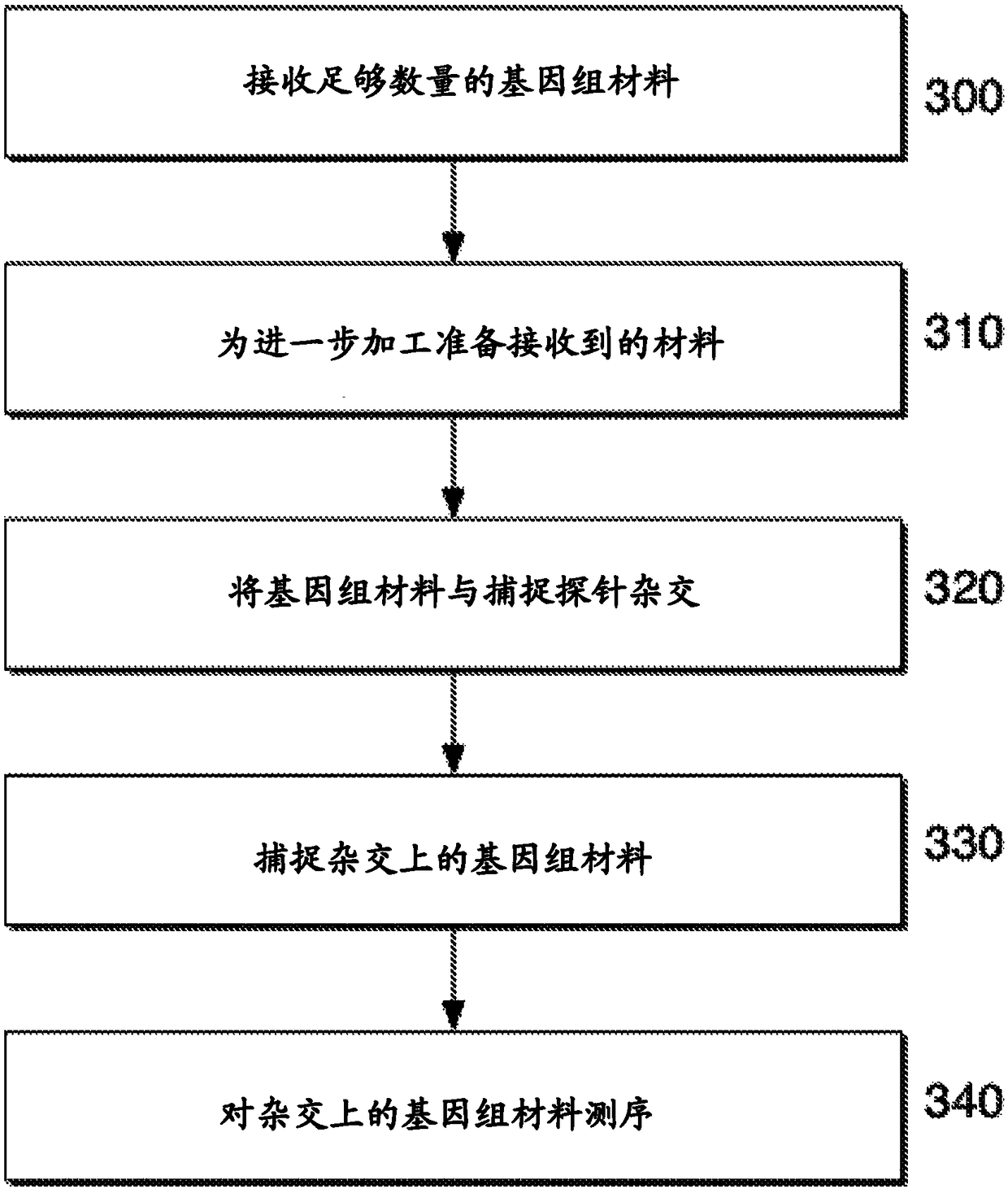
Deep sequencing profiling of tumors
A technology for sequencing and tumor, applied in the field of deep sequencing profiling analysis of tumors, which can solve problems such as elevation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0126] Example 1 - Sample library preparation using KAPA HyperPlus library preparation
[0127] 1.1. Resuspend lyophilized indexing adapters (Adapter Kit A).
[0128] 1.1.1. Spin down the lyophilized indexing adapters contained in SeqCap Adapter Kit A and / or B briefly to allow the contents to pellet at the bottom of the tube.
[0129] 1.1.2. Add 50 microliters of cold PCR grade water to each of the 12 tubes labeled 'SeqCap Index Adapter' in SeqCap Adapter Kit A and / or B. Keep adapters on ice.
[0130] 1.1.3. Briefly vortex the index adapters with PCR grade water and swirl down to resuspend the index adapter tubes.
[0131] 1.1.4. Tubes of resuspended index adapters should be stored at -15 to -25°C.
[0132] 1.2. Preparation of sample library
[0133] 1.2.1. Dilute the gDNA to be used for library construction (about 100 ng to about 1 microgram) in 10 mM Tris-HCl (pH 8.0) to a total volume of 35 microliters in a 0.2 ml tube or well of a PCR plate.
[0134] 1.2.2. Assemble...
Embodiment 2
[0178] Example 2 - Hybridization Sample and SeqCap EZ Probe Pool
[0179] 2.1.1. Allow AMPure XP Reagent to warm to room temperature for at least 30 minutes before use.
[0180] 2.1.2. Add 5 microliters of COT Human DNA (1 mg / ml) contained in the SeqCap EZ Accessory Kit v2 to a new tube / well.
[0181] 2.1.3. Add 3 μg DNA sample library to sample containing 5 μl COT human DNA. Multiple libraries constructed from the same sample can be combined for this purpose.
[0182] 2.1.4. Add 2,000 pmol (or 2 µl) of Hybridization Enhancer Oligomer (1 µl of 1,000 pmol SeqCap HE Universal Oligomer and 1 µl of 1,000 pmol SeqCap HE Indexing Oligomer matching the sample library adapter index) Add to DNA sample library plus COT human DNA.
[0183] 2.1.5. Determine the total volume of the above mixture by adding the input volumes of COT Human DNA, DNA Sample Library Pool, SeqCap HE Universal Oligomer and SeqCap HE Index Oligomer Pool.
[0184] 2.1.6. Add 2 volumes of AMPure XP Reagent (equi...
Embodiment 3
[0204] Example 3 - Cleaning and Recovery of Captured DNA Sample Libraries
[0205] 3.1. Dilute the 10X Wash Buffer (I, II, III and Stringent) and 2.5X Bead Wash Buffer contained in the SeqCap Hybridization and Wash Kit to create a 1X working solution. The volumes listed below are sufficient for one subcapture.
[0206]
[0207] 3.2. Preparation of capture beads
[0208] 3.2.1. Allow the capture beads contained in the SeqCap Pure Capture Bead Kit to equilibrate to room temperature for 30 minutes before use.
[0209] 3.2.2. Vortex the capture beads for 15 seconds prior to use to ensure a homogeneous mixture of beads.
[0210] 3.2.3. Aliquot 50 microliters of beads for each capture into 0.2ml or 1.5ml tubes (ie use 50 microliters of beads for one capture and 200 microliters of beads for four captures, etc.). Sufficient beads for two captures and twelve captures can be prepared in one 0.2 ml tube and 1.5 ml tube, respectively.
[0211]3.2.4. Place the tube on the magnetic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com