Application of pir-mmu-31018127 exosomes in tongue sole
A technology of semi-smooth tongue sole and exosomes, which is applied in the directions of biochemical equipment and methods, microbial determination/inspection, etc., to achieve the effect of reliable identification results
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
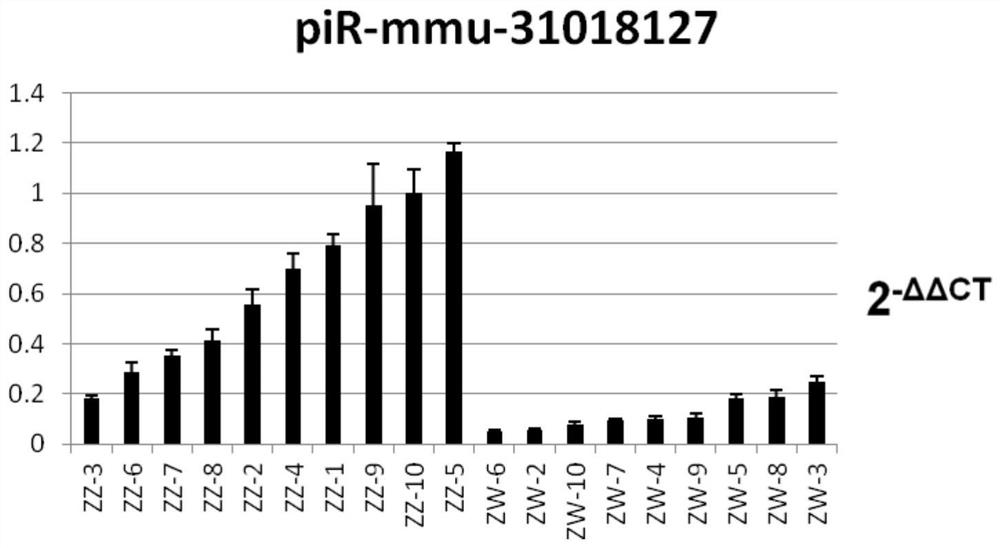
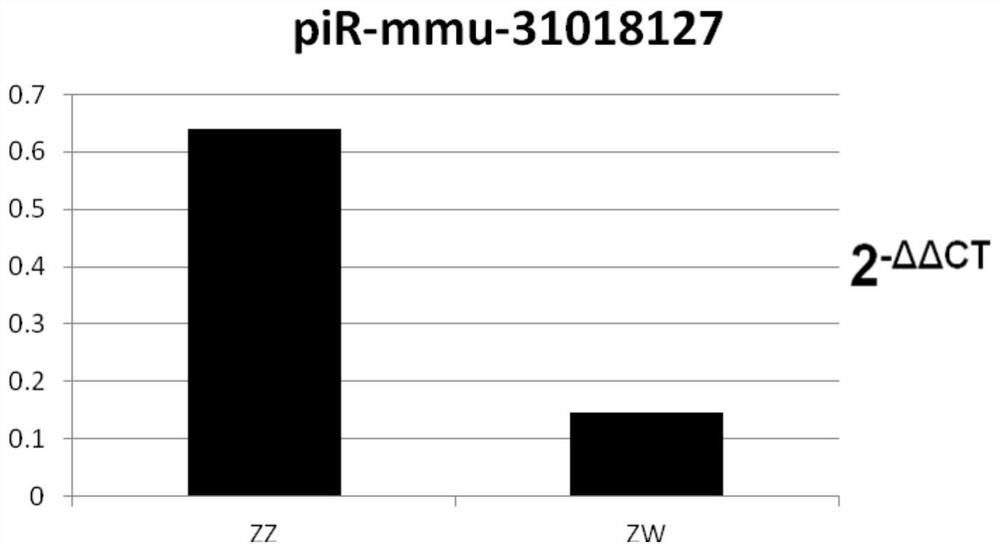
[0018] A screening method for exosomal piR-mmu-31018127 derived from tongue sole semen as a biomarker.
[0019] 1. Collection of semen samples
[0020] Collect 0.5 ml of semen from sexually mature males of semi-smooth tongue sole in a centrifuge tube for the isolation and identification of exosomes.
[0021] 2. Extraction of exosomes
[0022] (1) Transfer 0.5ml semen sample to 1.5ml EP tube, place at 4°C, centrifuge at 1200g for 15min to remove sperm cells, centrifuge at 15000g at 4°C for 20min to remove small cell impurities and debris, dilute 1 times with PBS, and use 0.45um filter membrane Perform pre-filtration, and then filter the filtrate through a 0.22um membrane filter.
[0023] (2) Exosomes were extracted from the filtered samples using the Total Exosome Isolation Kit.
[0024] (3) Collect the lysate and transfer completely to an RNase-free 2ml tube.
[0025] 3. Identification of exosomes: Hitachi H600IV transmission electron microscope observation, after transmis...
Embodiment 2
[0031] Kit for identification of male and pseudo-male by exosome miRNA markers in semismooth tongue sole semen
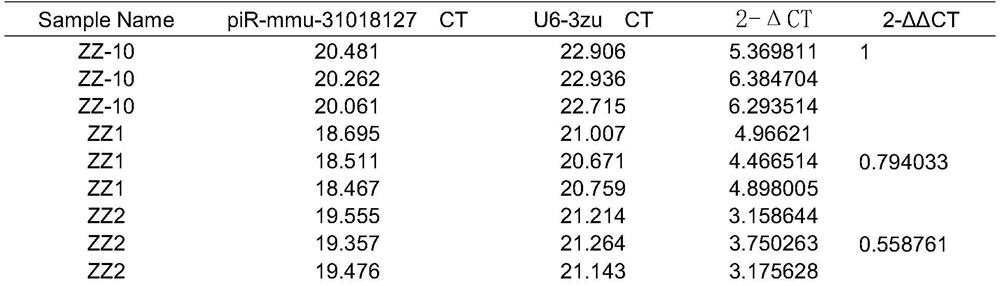
[0032] The identification kit based on semen exosome piRNA marker piR-mmu-31018127 includes piR-mmu-31018127 quantitative detection primers, primers, F:TACGCATGTGGTTCAGTGGT, R:TCGTATCCAGTGCAGGGTC, reverse transcription primer is piR-mmu-31018127-RT: GTCGTATCCAGTGCAGGGTCCGAGGTATTCGCACACTGGCGAGACG , and other PCR reverse transcription reagents and PCR fluorescent quantitative reagents, the internal reference piR is U6, and the primers are U6-F: CTCGCTTCGGCAGCACATACT; U6-R: ACGCTTCACGAATTTGCGTGTC; primers include reverse transcription primers, quantitative PCR forward primers and reverse primers, and other reagents The box contains other conventional reagents for quantitative PCR reactions: reverse transcriptase, Taq enzyme, dNTP, buffer, Mgcl 2 , DEPC water and reference substance. The reaction system used in this kit is a 10ul system, that is, 0.5ul 10*miRNA primer ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com