Nucleic acid reagents, kits and systems for detecting enteroviruses
An enterovirus and kit technology, which is applied in the field of enterovirus detection, can solve the problems of complex sample processing, increase operational complexity, and judge complex results, achieve simple detection process, avoid sample extraction steps, and automatic results. interpret and effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
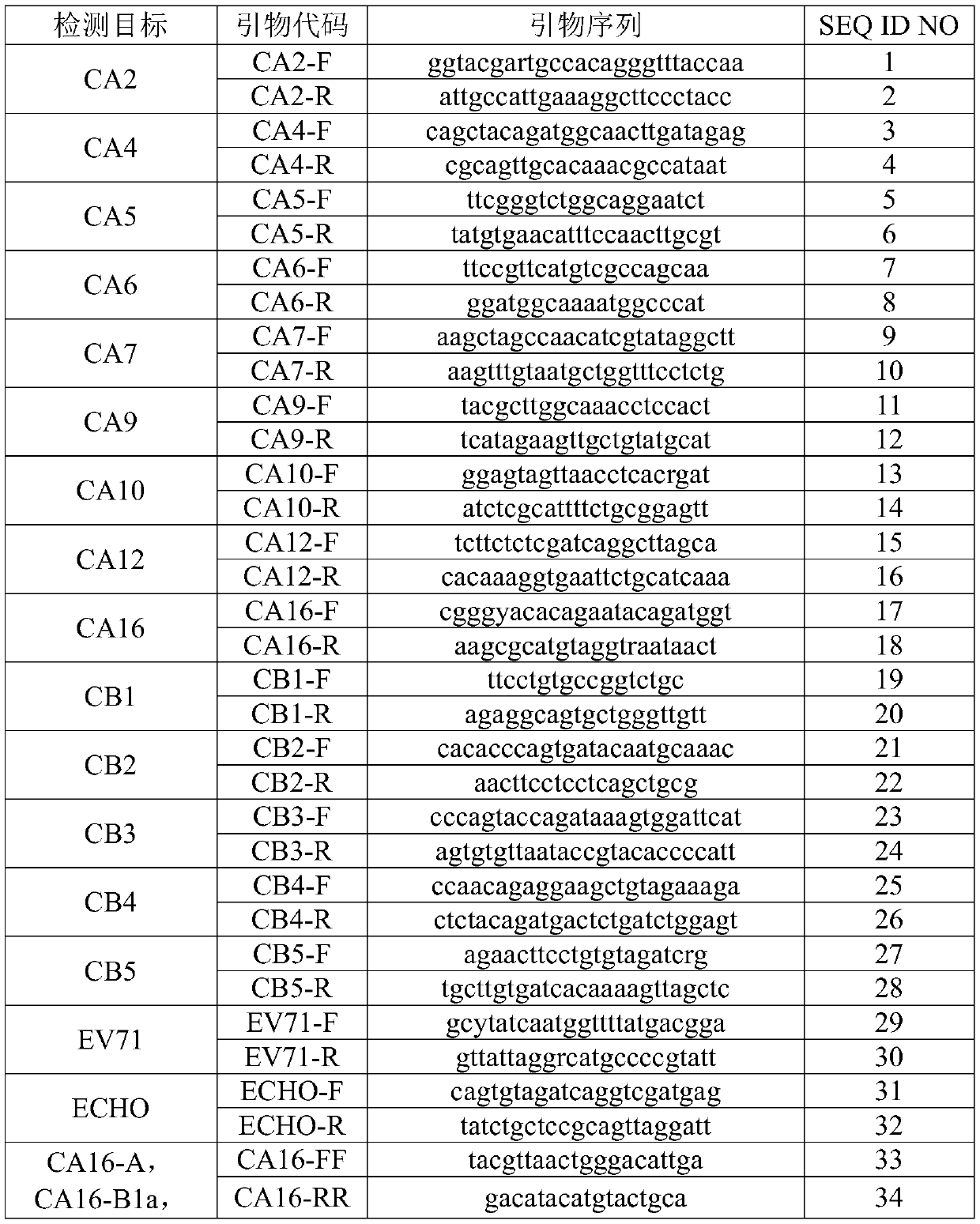
[0048] 1. Primer and probe synthesis
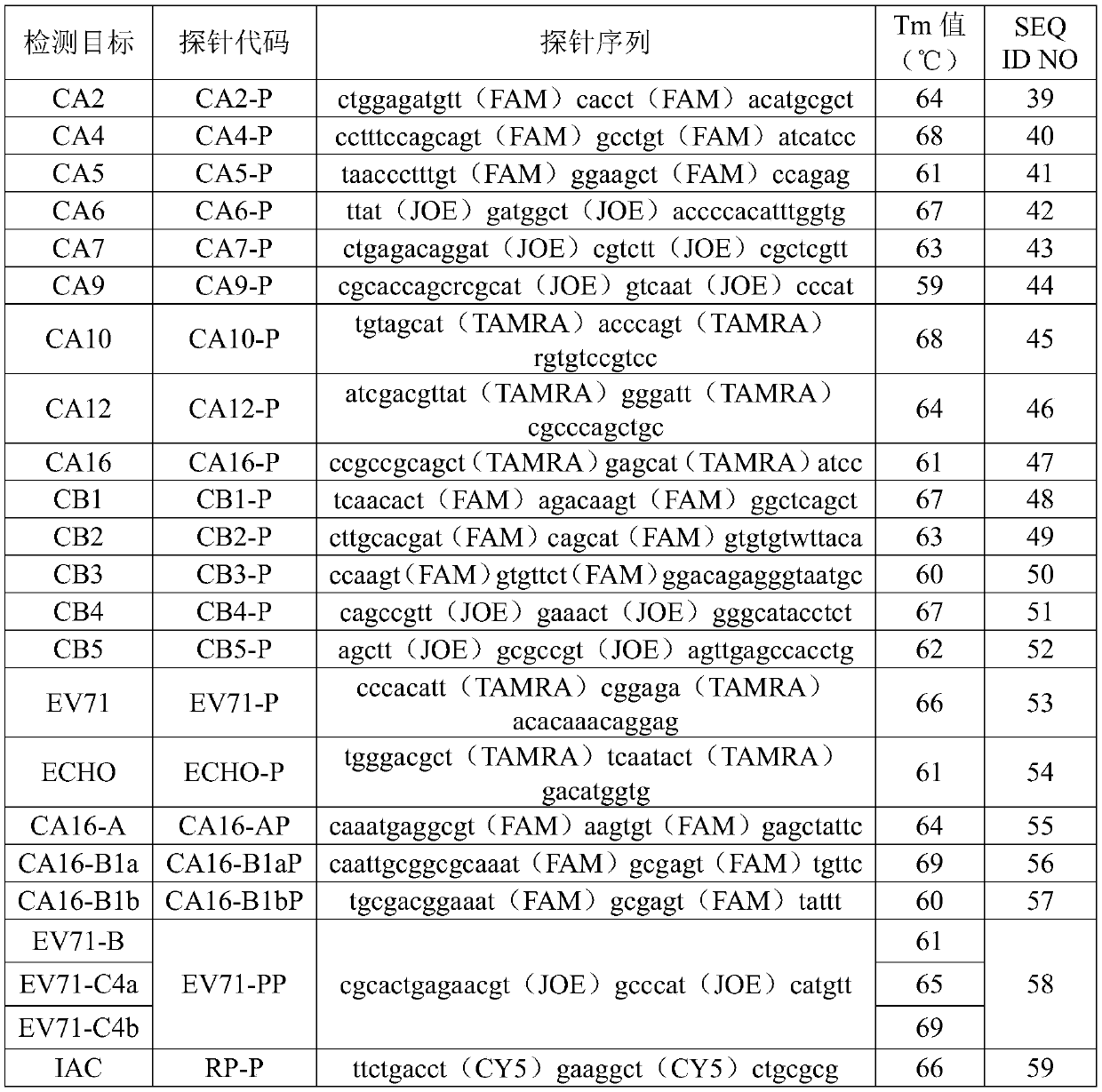
[0049] Sequence synthesis was performed according to the primer and probe sequences shown in Table 1 and Table 2. In the sequence, Y represents the degenerate base T / C; R represents the degenerate base A / G; W represents the degenerate base A / T; FAM in the probe is 6-carboxyfluorescein, JOE is 2,7-di Methyl-4,5-dichloro-6-carboxyfluorescein, TAMRA is 6-carboxytetramethylrhodamine, CY5 is 5H-indocyanine. The brackets in the probe sequences in Table 2 indicate that the t on the left side of the brackets has a fluorescent label, and the contents in the brackets indicate the choice of fluorescent labels.
[0050] Table 1
[0051]
[0052]
[0053] Table 2
[0054]
[0055] 2. Template extraction
[0056]After collecting clinical samples such as nasopharyngeal swabs with the sampler matched with ParaDNA, they can be amplified directly in the ParaDNA reactor.
[0057] 3. Construct Hybeacon probe technology detection system
[0058...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com