Reduced memory nucleotide sequence comparison
A nucleotide sequence, nucleotide technology, applied in sequence analysis, instruments, complex mathematical operations, etc., can solve problems such as slow memory
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
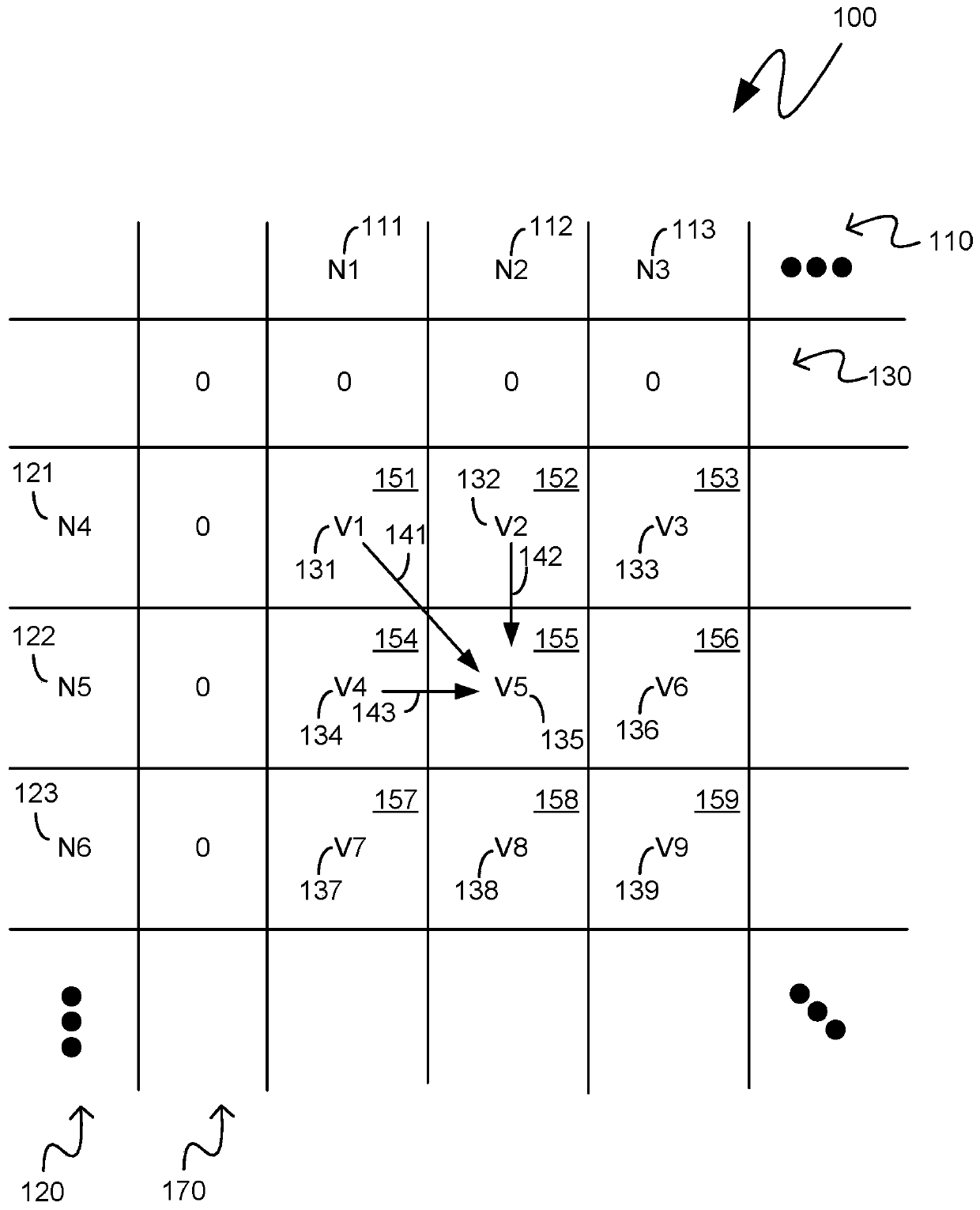
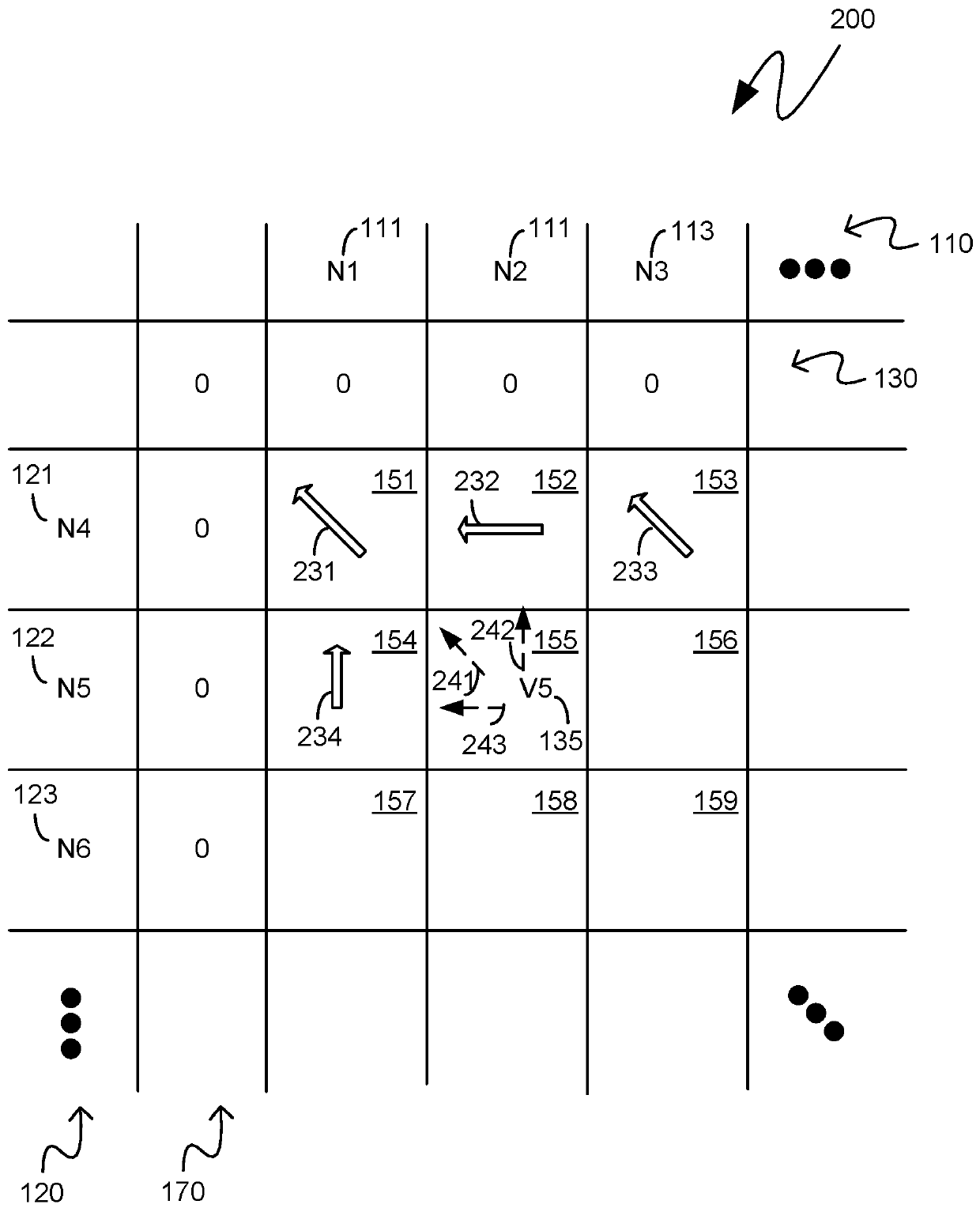
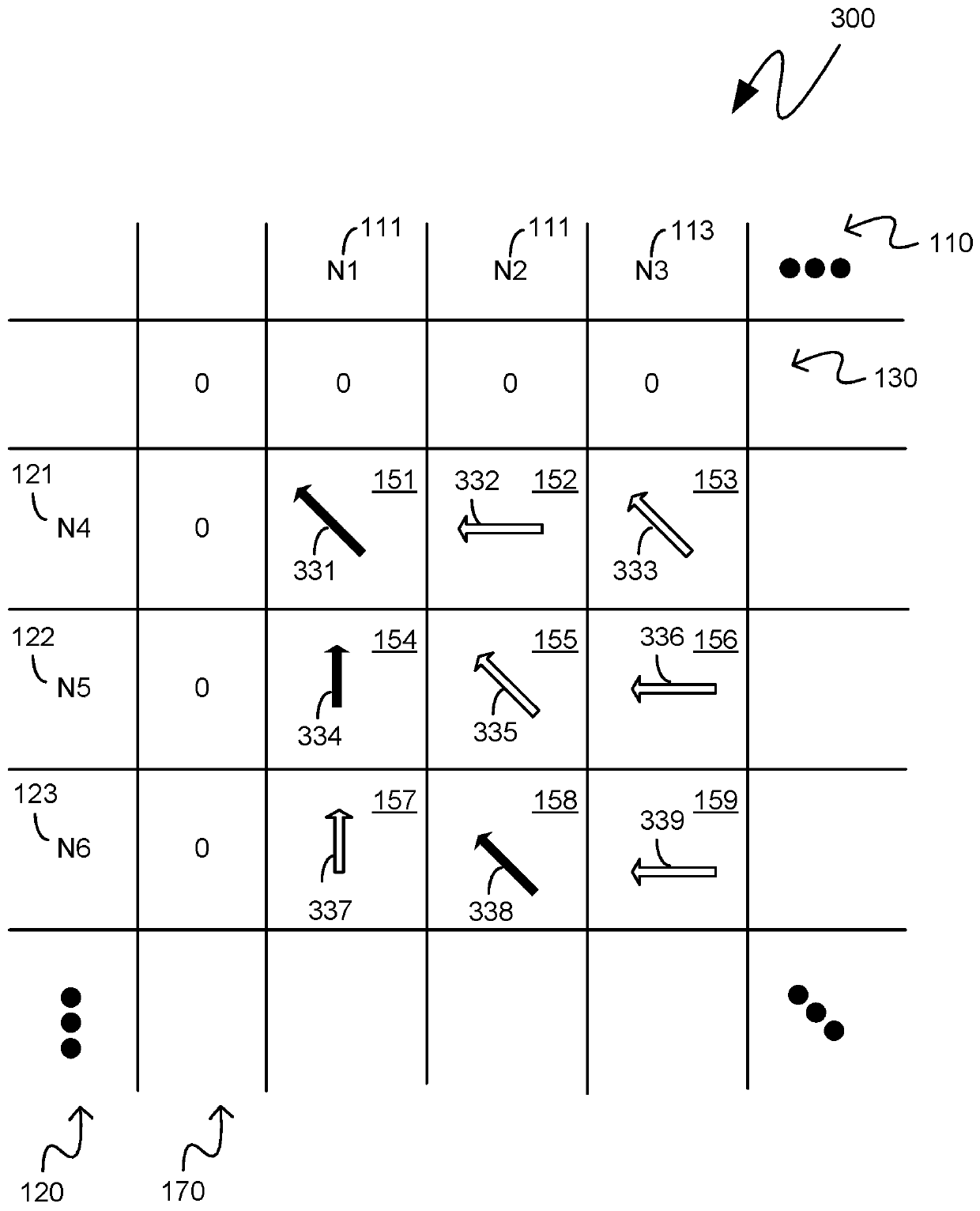
[0015] The following description pertains to custom integrated circuit systems that operate at a reduced memory footprint by storing and referencing only a single portion, or segment, of a two-dimensional matrix representing a comparison between two nucleotide sequences. uses Smith Waterman analysis to compare two strings of nucleotide sequences. The first segment of the two-dimensional matrix initially retained in memory may be a segment comprising cells from which backtracking may be performed for the purpose of generating a text string comprising two nucleotide sequences indicators of similarity and difference. As the backtracking proceeds, there may be a need to backtrack to metadata corresponding to cells from segments that are not currently held in memory. Such a segment may be regenerated from previously generated scores associated with a checkpoint cell of the two-dimensional matrix comprising both edges of the regenerated segment. In this way, backtracking can be pe...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com