A base editor and its preparation method and application
An editor and gene editing technology, applied in the field of gene editing, can solve problems such as narrow application range and rare targets
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0029]The invention provides the preparation method of the base editor, comprising the following steps: inserting the DNA fragment of the dCpf1-RR-eBE expression frame into the vector backbone pCMV-dCpf1-eBE to construct and obtain the pCMV-dCpf1-RR-eBE recombinant plasmid ; The sgRNA general expression frame DNA fragment was inserted into the vector backbone pUC57 to obtain the pLbCpf1-sgRNA recombinant plasmid.
[0030] In the present invention, the insertion site of the dCpf1-RR-eBE expression cassette DNA fragment is between the Pst I restriction site and the Apa I restriction site of the vector backbone pCMV-dCpf1-eBE, that is, the vector backbone The 2365bp-5178bp interval of pCMV-dCpf1-eBE; in the present invention, the insertion is preferably performed by performing double enzyme digestion on the dCpf1-RR-eBE expression cassette DNA fragment and pCMV-dCpf1-eBE respectively; The enzymes used for double enzyme cutting are Pst I enzyme and Apa I enzyme. In the present in...
Embodiment 1
[0040] Construction of base editors
[0041] 1. Construction of pCMV-dCpf1-RR-eBE recombinant plasmid
[0042] The DNA fragment of the dCpf1-RR-eBE expression cassette of 2814bp was synthesized (the nucleotide sequence is shown in SEQ ID NO: 1), and inserted into the pCMV-dCpf1-eBE vector by double digestion with Pst I enzyme and Apa I enzyme to obtain pCMV - dCpf1-RR-eBE vector.
[0043] The pCMV-dCpf1-eBE vector was purchased from addgene, Cat. No. 107688.
[0044] Pst I enzyme was purchased from Treasure Bioengineering (Dalian) Co., Ltd., product number 1624; ApaI enzyme was purchased from Treasure Bioengineering (Dalian) Co., Ltd., product number 1604.
[0045] Enzyme digestion system: 50 μL, reagents purchased from Treasure Bioengineering (Dalian) Co., Ltd.): Pst I enzyme 1 μL, Apa I enzyme 1 μL, dCpf1-RR-eBE expression cassette DNA fragment or pCMV-dCpf1-eBE backbone vector 1 μg, BufferH 5 μL , add double distilled water to 50 μL. The digestion temperature is 37°C, a...
Embodiment 2
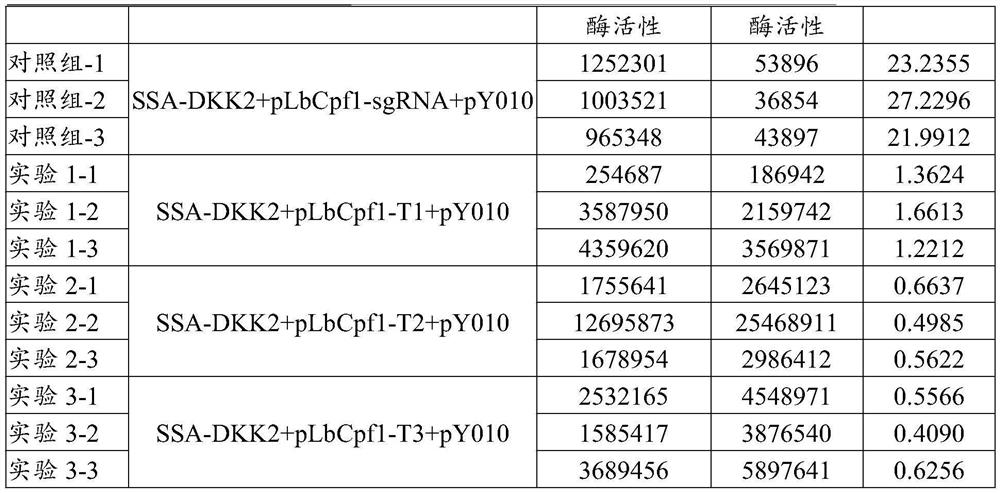
[0070] Application of base editors in gene editing of mammalian cell lines
[0071] Diqing sheep skin epithelial cell line DQSHS1 was purchased from the Kunming Cell Bank of the Chinese Academy of Sciences, number: KCB 94026.
[0072] 1. sgRNA target design
[0073] The sequence of the sheep DKK2 gene including the first exon (DKK2-440, shown below) was extracted from the sequence of sheep chromosome 6 (NCBI GI:417531944), and the Cpf1 sgRNA target was designed.
[0074] agactgagttcacacggtgctgggcccccaaagc caagtg gggttgggggaacagagtctgcgagtcccggcgccccgagt gcagggccccgtgttggggtcctccttcccatttgt atccgt atccttgcgggctttgcgcctccccgggggaccccctcgccgggagatggccgcactgatgcggggcaaggactcctcccgctgcctgctcctactggccgcggtgctgatggtggagagctcacagttcggcagct c gcgggc caaactcaactccatcaagtcctctctgggcggggagacgcctgcccaggccgccaatcgatctgcgggcacttaccaag gactggctttcggcggcagtaagaagggcaaaaacctggggcaggtaggaaaatacccccaatacactcttcaaccagaagaggtag ggacccg (SEQ ID NO: 12)
[0075] Target site TF3: a tccgt (SEQ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com