A kind of antigen for detecting invasive candidiasis albicans and use thereof
A kind of Candida albicans, invasive technology, applied in the field of biomedicine, can solve the difficult and accurate diagnosis of invasive Candida albicans infection and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0014] Embodiment 1 sequence selection
[0015] Using NCBI ( http: / / www.ncbi.nlm.nih.gov ) integrated protein search function, using "HWP1" as the search field, searched for the amino acid sequence of hyphal wall protein 1 (Candida albicans), excluded interference items, and selected HWP1 with GenBank accession No. AAC96368 as the analysis sequence (the full length of the sequence is 634aa), The sequence is shown as SEQ ID NO.1:
[0016] Using the NCBI amino acid sequence BLAST function, the selected sequences were compared online.
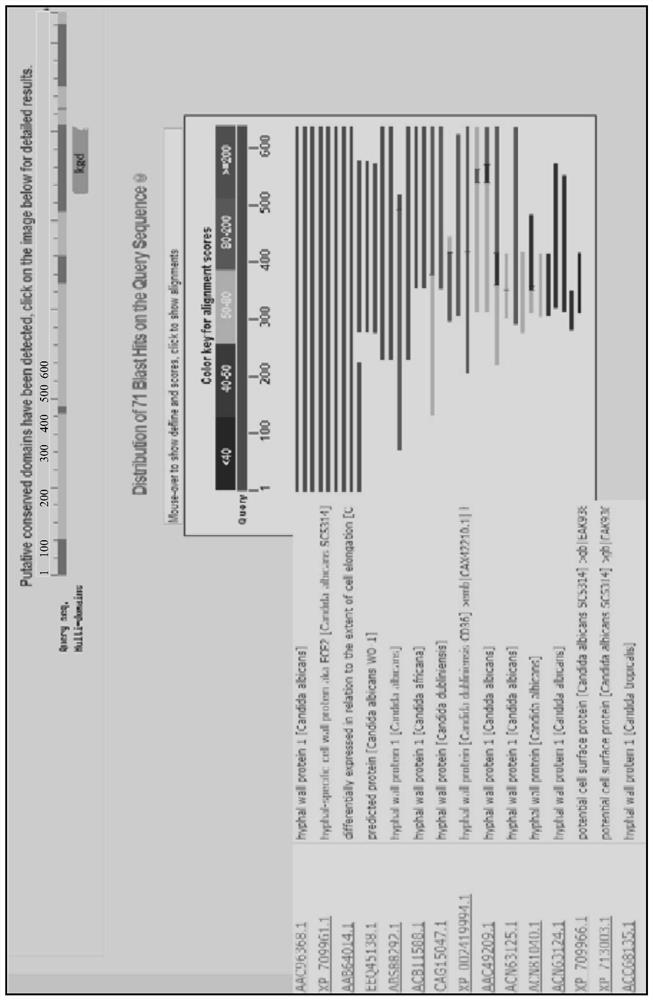
[0017] Such as figure 1 Sequence comparison results show that: the query sequence has 100% homology with the complete sequence included in NCBI, and also has 100% homology with the included non-full-length sequences within their respective lengths, indicating that the selected HWP1 protein primary structure amino acid Sequences are representative and sequences are available.
Embodiment 2B
[0018] Example 2 B cell epitope prediction
[0019] Comprehensive comparison of multiple antigen prediction analysis software [Godkin A J, Smith KJ, Willis A, et al. Naturally processed HLA class II peptides reveal highly conserved immunogenic flanking region sequence preferences that reflect antinprocessing rather than peptide-MHC interactions. J Immumol, 2001, 166( 11):6720-6727][Nielsen M, Lund O.NN-align-A neural network-based alignment algorithm for MHC class II peptide binding prediction.BMC Bioinformatics.2009,18;10:296.][Nielsen M,Lundegaard C , Lund O. Prediction of MHC class II binding affinity using SMM-align, a novel stabilization matrix alignment method. BMC Bioinformatics. 2007, 8:238.].
[0020] B cell epitope prediction adopts web server PREDICTED ANTIGENIC PEPTIDES, Bcepred( http: / / www.imtech.res.in / raghava / bcipep ), the parameters are set as follows: Immunogenicity="Immunodominant"; Mesure of Neutralization="YES"; Pathogen Group="Fungi" /
[0021] After a c...
Embodiment 3
[0024]Example 3 Th cell epitope prediction
[0025] MHC class II molecule Th antigen epitope prediction adopts the network server NetMHCII( http: / / www.cbs.dtu.dk / services / NetMHCII ) for online analysis [Saha.S, Raghava GP.BcePred: Prediction of Continuous B-Cell Epitopes in Antigenic Sequences Using recurrent neural network. Proteins. 2006, 65(1): 40-48.].
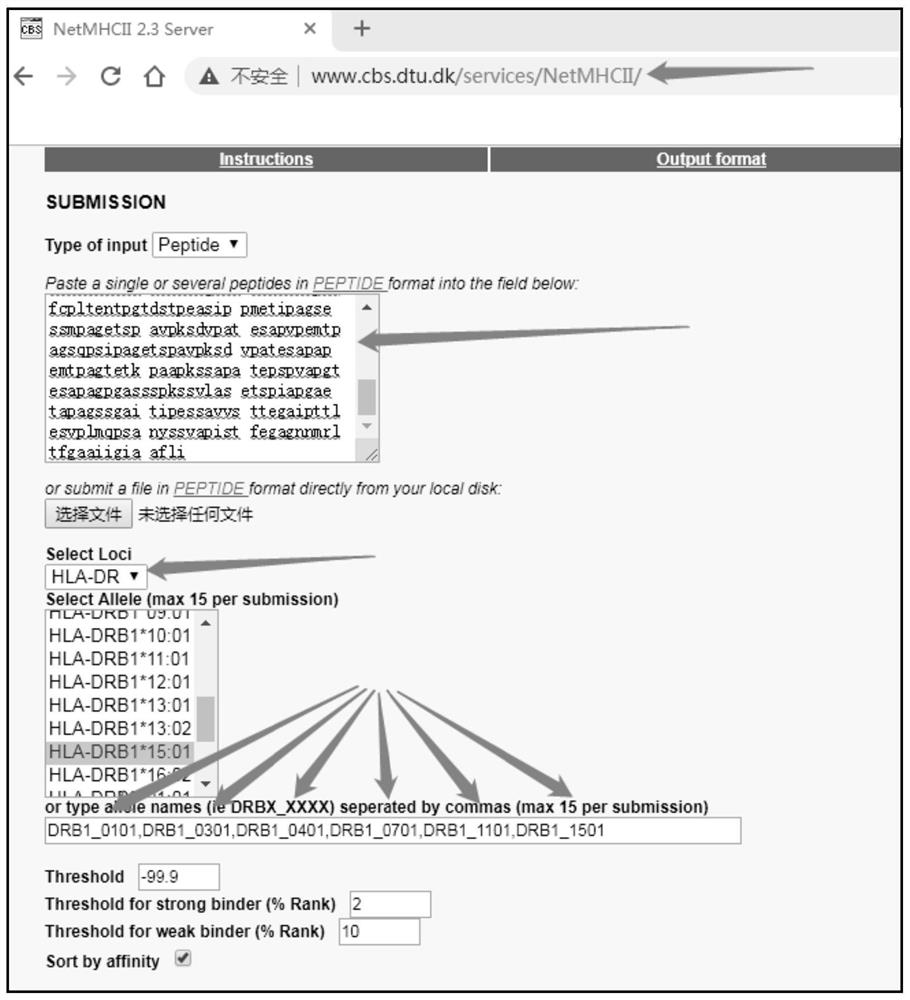
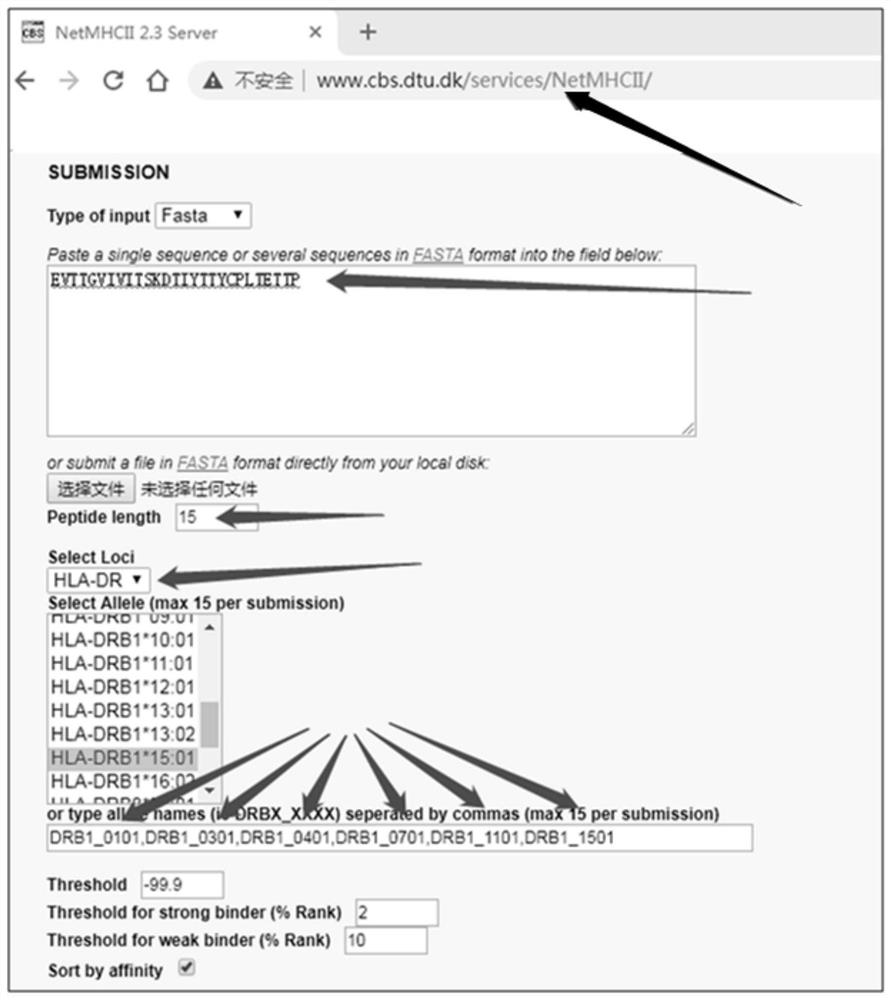
[0026] Select six epitopes HLA-DRB1*0101, HLA-DRB1*0301, HLA-DRB1*0401, HLA-DRB1* on the human leukocyte antigen (human leukocyte antigen, HLA) DR loci, which are widely representative 0701, HLA-DRB1*1101, and HLA-DRB1*1501 are used as binding sites, and every 15 amino acid residues are used as walking units to submit query tasks, download the Ecexl worksheet, analyze and verify that it may become a wide range of MHC II antigens HWP1 amino acid sequence of the epitope. Diagram of parameter settings for online prediction of Th cell epitope figure 2 , image 3 shown.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com