Adenine base editing tool and application thereof
A technology of adenine deaminase and purpose, applied in the direction of targeting specific cell fusion, introducing foreign genetic material using vectors, antibody mimics/scaffolds, etc., can solve problems such as limiting the application of base editors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
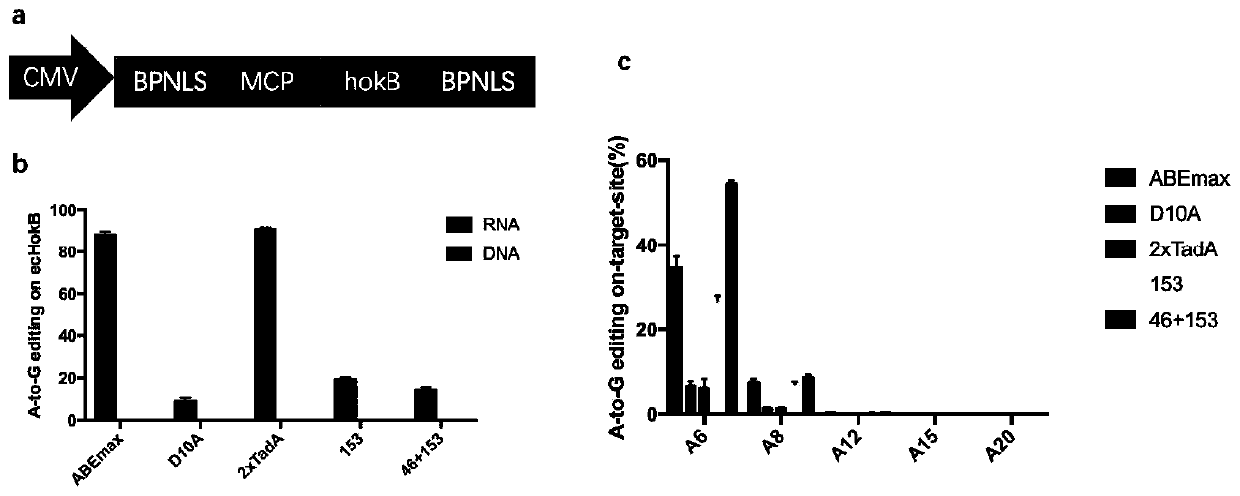
[0061] In this example, the hokB overexpression reporter system was used to verify the editing ability of eABEmax and the RNA off-target effect on HEK293T. The results are as follows figure 2 Shown.
[0062] 1.1 HokB overexpression reporter system plasmid construction
[0063] The hokB overexpression vector was constructed by Mut Express II Fast Mutagenesis Kit V2 (Vazyme, C214-02). The constructed hokB overexpression reporter system plasmid sequence is shown in SEQ ID NO.2.
[0064] 1.2 Construction of sgRNA plasmid
[0065] Design sgRNA and synthesize oligos, the upstream sequence is: 5'-accgGGCCCAGACTGAGCACGTGA-3' (SEQ ID NO. 3), the downstream sequence is: 5'-aaacTCACGTGCTCAGTCTGGGCC-3' (SEQ ID NO. 4), the upstream and downstream sequences are through the program (95 ℃, 5min; 95℃-85℃at-2℃ / s; 85℃-25℃at-0.1℃ / s; hold at 4℃) Annealing, connect to pGL3-U6 linearized by BsaI (NEB: R0539L) -sgRNA (Addgene#51133) vector. The linearization system is as follows: pGL3-U6-sgRNA 2μg; buffe...
Embodiment 2
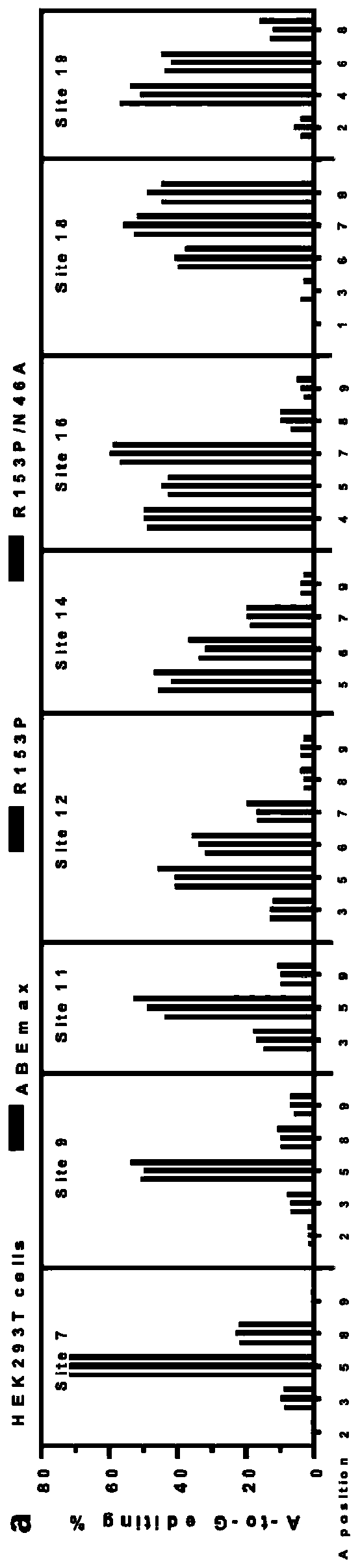
[0073] In this example, eABEmax was used to edit endogenous gene loci on HEK293T cells.
[0074] 2.1 Construction of sgRNA plasmid
[0075] Eight human endogenous base sites were selected and sgRNA was designed. The polynucleotide sequences of oligos used are shown in SEQ ID NOs. 9 to 26. The positions of the eight sgRNAs used in the genome are NC_000004.12:52670026- 52670045; NC_000022.11: 19401208-19401227; NC_000015.10: 47301029-47301048; NC_000014.9: 88099864-88099883; NC_000001.11: 154311091-154311110; NC_000001.11: 179826686-179826705; NC_000001.11: 184974900-184974919; NC_000001.11: 184974901-184974920. Follow 1.2 to construct sgRNA plasmid.
[0076] 2.2 Cell culture, transfection and identification
[0077] HEK293T cells were cultured and transfected according to 1.3. The amount of transfected plasmid was 0.6 g of eABEmax and 0.3 g of sgRNA expression vector plasmid. ABEmax was used as a control. After 12 hours of transfection, puromycin was added according to the above 1.3...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com