Primer group and kit for detecting mutant and deletion genes of thalassemia and application of primer group and kit
A technology for thalassemia and gene detection, which is applied in the direction of recombinant DNA technology, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of inability to find mutation sites, increase reagent costs, and high costs, and avoid secondary Effects of amplification and purification, increased reagent cost, and reduced reagent cost
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] The kit of the invention is used to detect known genotype samples by using multiplex PCR+nested PCR technology.
[0034] 1. Primer Design
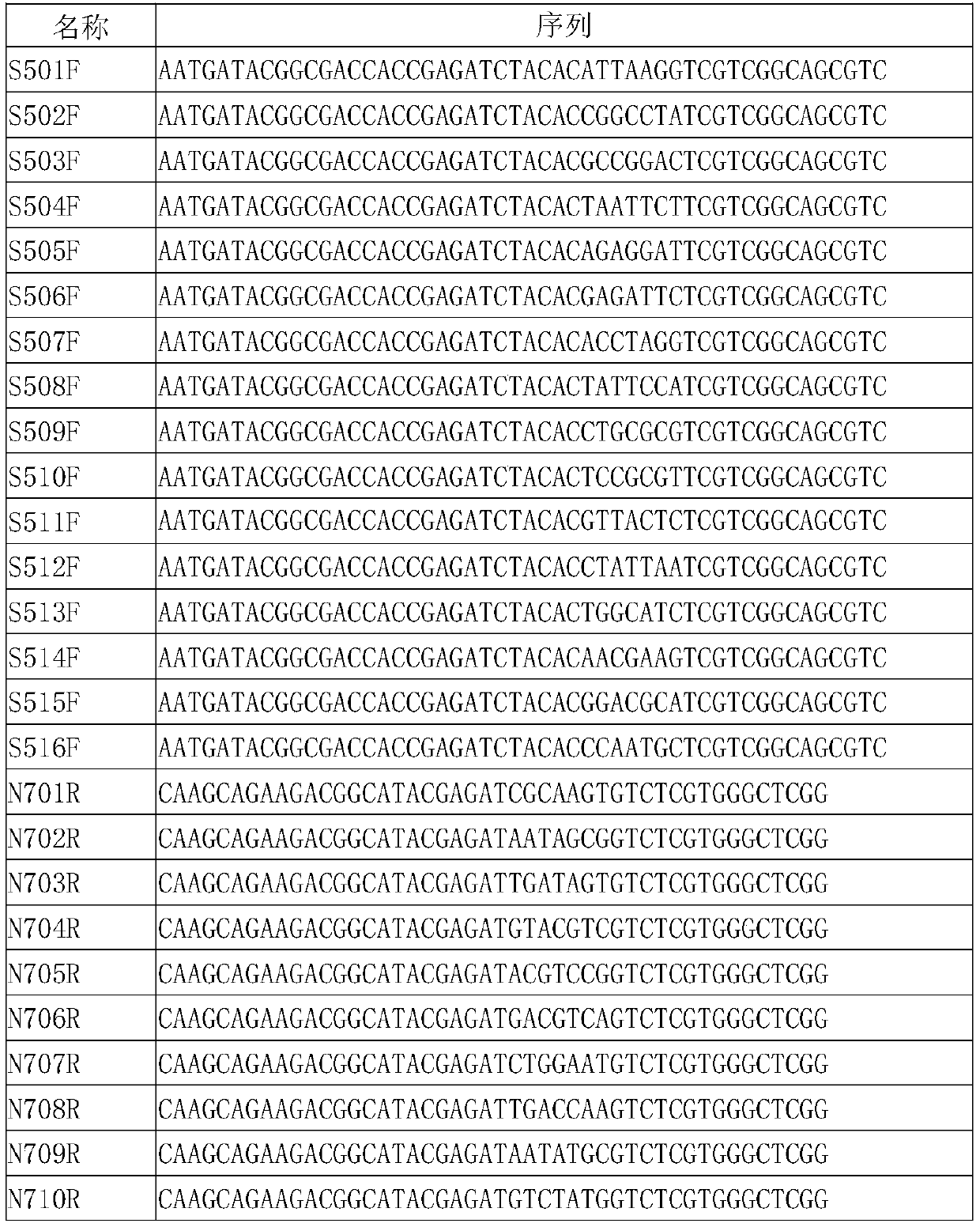
[0035]First, conduct bioinformatics analysis on HBA1, HBA2, and HBB gene sequences to find out the conserved and specific sequences of HBA1, HBA2, and HBB genes. Know the intron mutation position to design primers. Next, use the known mutant samples as test samples, use the screened secondary candidate primer pairs for PCR amplification, and perform Sanger sequencing on the PCR products. The sequencing results are compared with the reference gene sequence. Alignment, the sequences are consistent and the quality of the sequencing peak is good, as the final primer pair, the primer sequence is SEQ ID1-SEQ ID32, and the amplification product is about 200-300bp. The primer list is as follows:
[0036] name Primer sequence name Primer sequence SEQ-1 CAGACAGGGAGGGGAAATGA SEQ-17 AAATGCACTGACCTCCCCACA SEQ-2 ...
Embodiment 2
[0074] Using the kit of the present invention, multiplex PCR+nested PCR technology is used to detect clinical samples. The clinical samples are derived from samples whose genotypes have been determined by conventional PCR-RDB technology. The genotypes determined in this sample group include: SEA Deletion, 3.7 deletion, 4.2 deletion, QS mutation, CS mutation, WS mutation, CD41-42, IVS-II-654, CD17, -28, βE, CD71-72, IVS-1-1, CD43, CD27-28.
[0075] 1. Primer Design
[0076] The design of primers is the same as in Example 1
[0077] 2. DNA extraction and primer set preparation
[0078] 1) Sample DNA extraction
[0079] Nucleic acid was extracted from clinical samples using conventional DNA extraction reagents to obtain DNA samples from clinical samples of α-thalassemia to be tested.
[0080] Clinical test samples were collected from Nanning Maternal and Child Health Hospital (2000 cases, 715 cases of deletion type, 631 cases of mutant thalassemia samples, 654 cases of normal ...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap