Drug protein binding rate prediction method and system based on structure and grade classification
A technology of protein binding rate and grade classification, which is applied in the field of prediction of drug protein binding rate of grade classification, can solve the problem of low prediction accuracy of high binding drugs, achieve the effect of reducing research and development risks, improving prediction accuracy, and increasing applicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
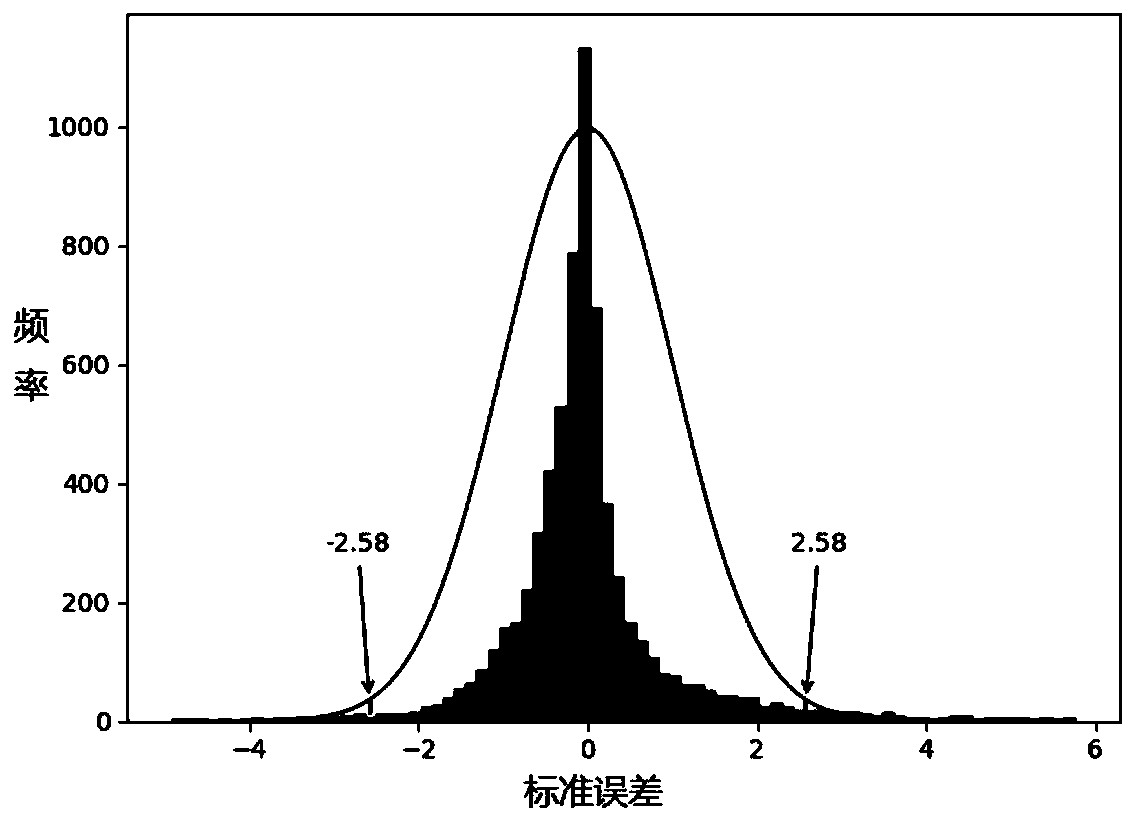
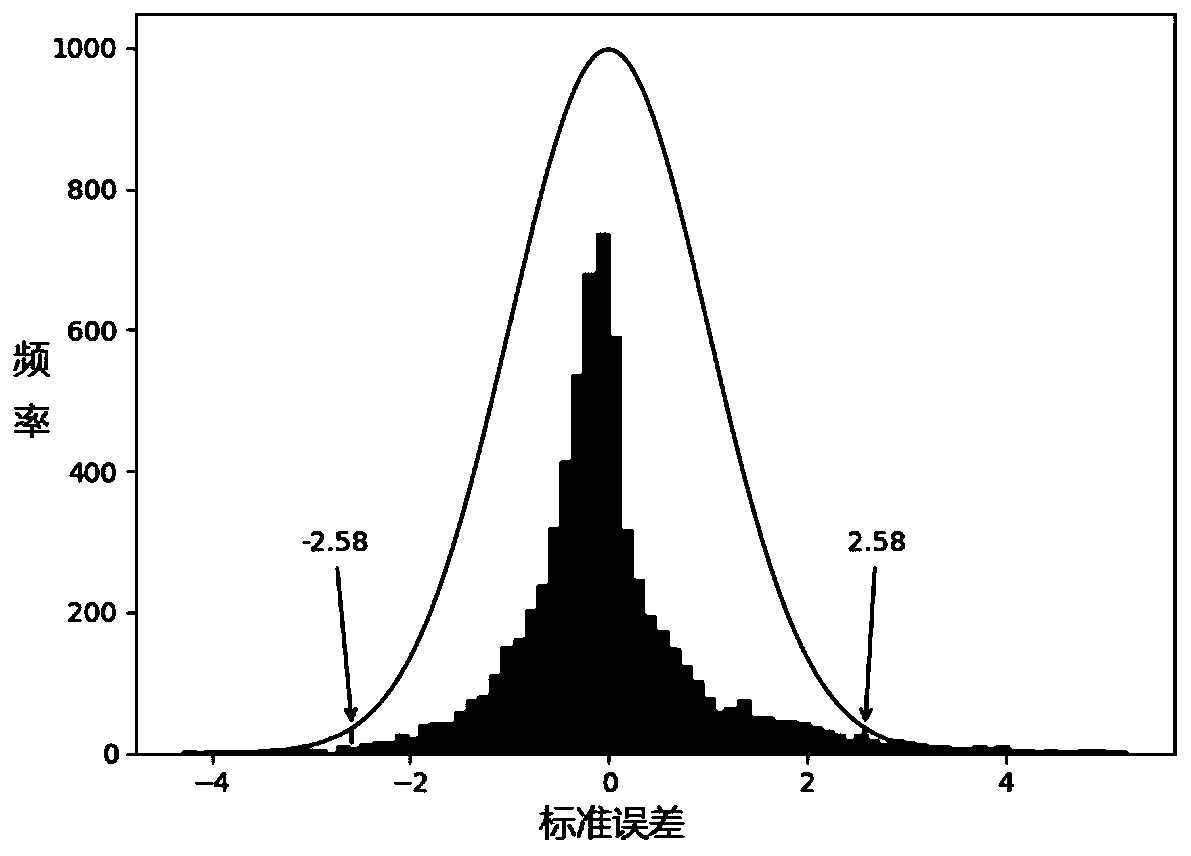
Image
Examples
Embodiment
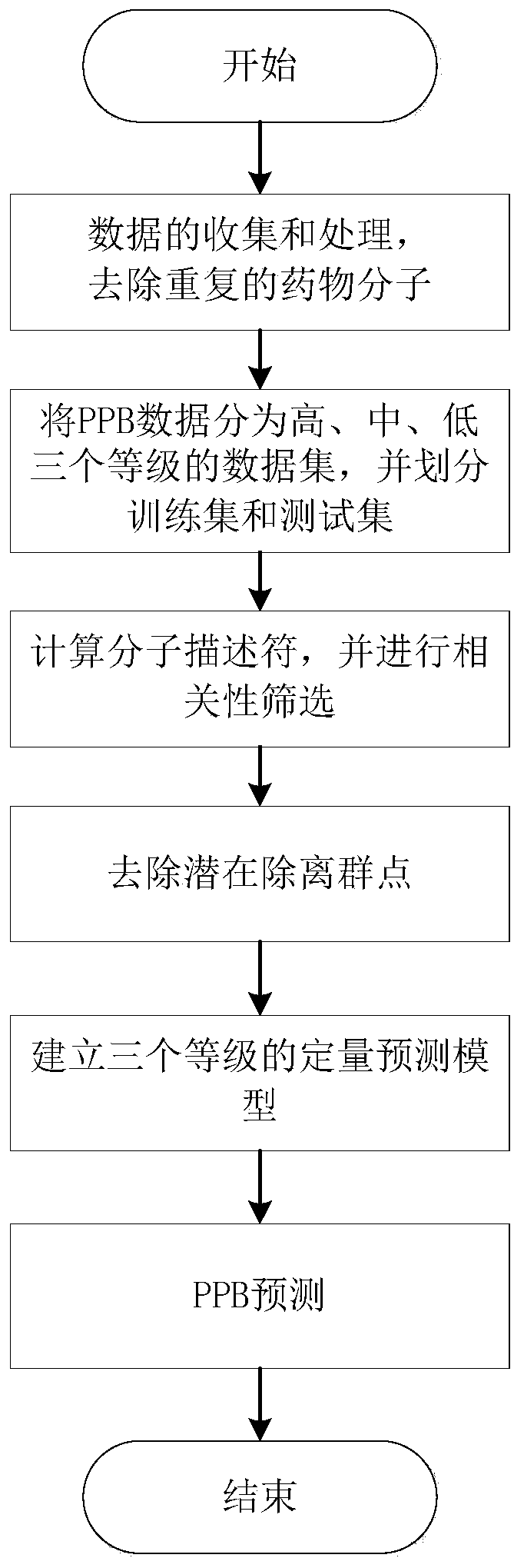
[0040] This embodiment provides a method for predicting drug protein binding rate based on structure and rank classification, such as figure 1 shown, including:
[0041] Step 1, data collection and processing:
[0042] (1) By consulting literature and databases, a total of 12,646 protein binding rate (PPB) data values and corresponding structural codes of drug molecules were collected.
[0043] (2) Process the collected data:
[0044] If the collected PPB value is within a numerical range (for example: 0.96~0.97), take the mean value of the numerical range as the PPB value of the drug molecule; if the collected PPB value is greater than or less than a certain value (for example: >0.99) , if there is a more reliable data source, select the PPB value from a more reliable source as the PPB value of the drug molecule, if not, take the fixed value 0.99 as the PPB value of the drug molecule.
[0045] According to the naming, structure coding and properties of the drug molecule,...
PUM
 Login to view more
Login to view more Abstract
Description
Claims
Application Information
 Login to view more
Login to view more - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap