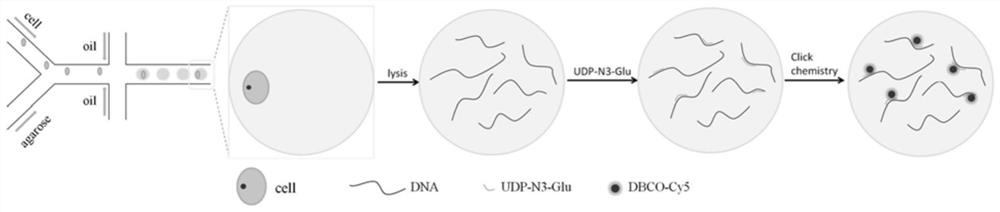
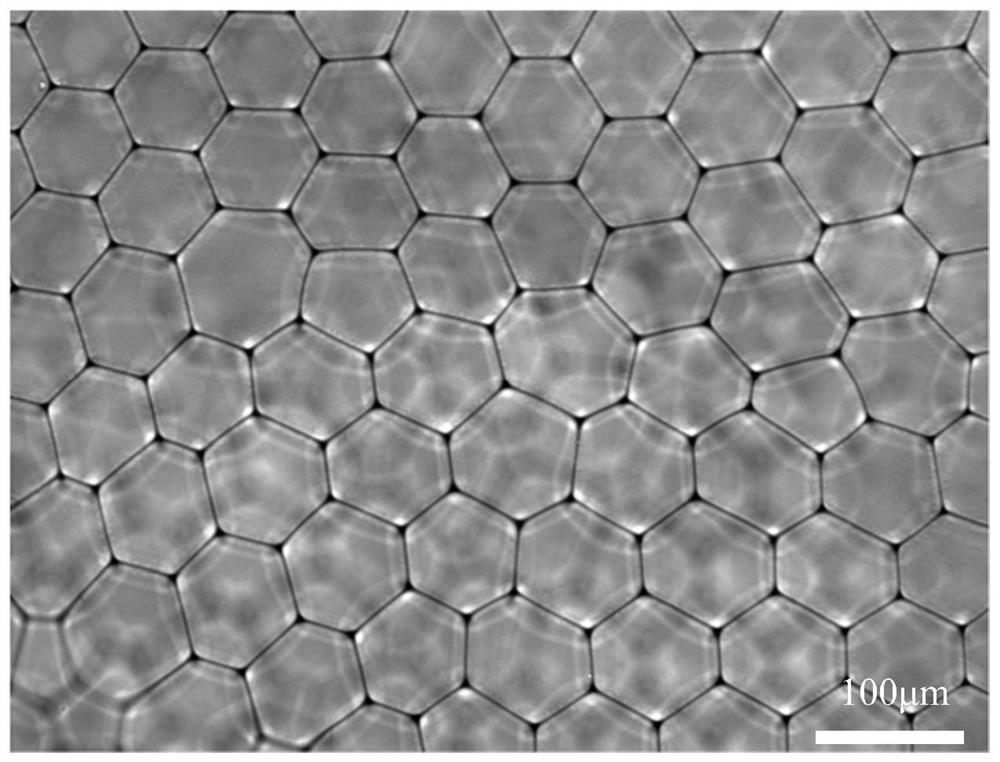
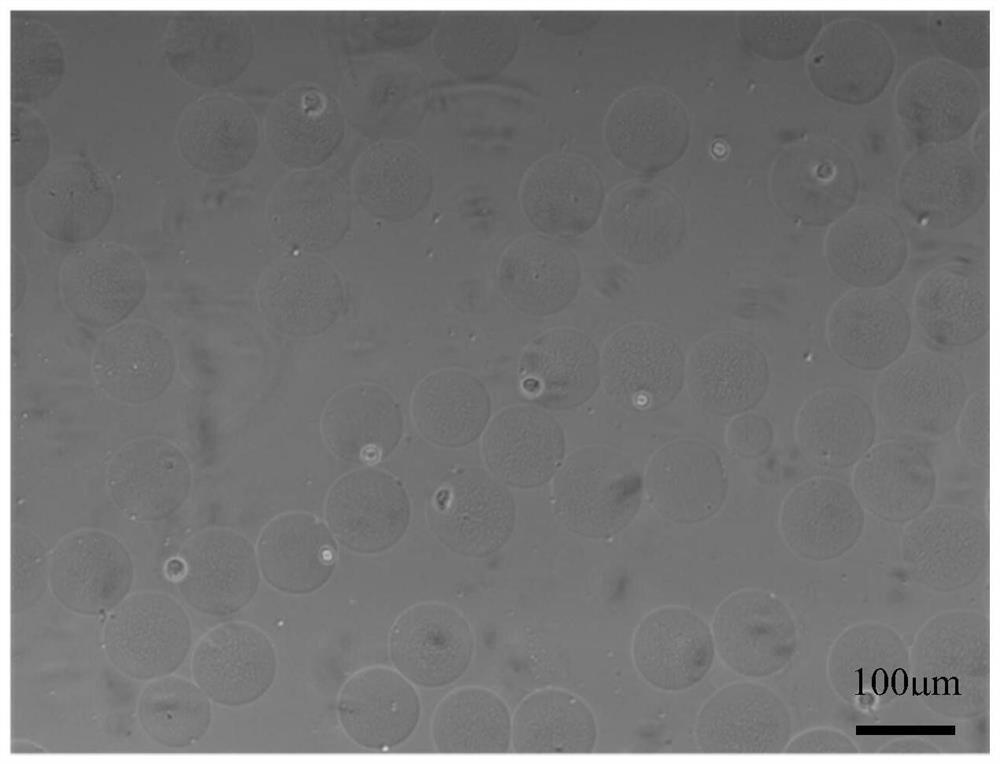
A method for detecting single cell 5-hmc
A detection method and single-cell technology, which is applied in the detection of modified bases and the field of analytical chemistry, can solve the problems of harsh chemical reaction conditions and low reaction efficiency, and achieve improved accuracy and sensitivity, simple reaction conditions, accurate detection and analysis Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0042] Centrifuge the solidified single-cell gel microspheres at 3000rr for 2min and remove the supernatant; add isopropanol to the gelatin, shake and mix evenly, and centrifuge at 3000r for 2min to remove the supernatant; add absolute ethanol to the gelatin, After oscillating and mixing evenly, centrifuge at 3000r for 2min, remove the supernatant, and repeat this step twice; add PBS to the rubber balls, shake and mix evenly, and centrifuge at 3000r for 2min, remove the supernatant; complete the deemulsification of the rubber balls. Add 50μl 2×SSC to the lysed rubber balls, vortex and mix well, then centrifuge at 3000r for 2min, remove the supernatant; add 20μM SYBR-Green to the rubber balls, and shake in the DEPC water reaction system at 37°C and 500rpm in a dark environment React for 1 hour; after the reaction, centrifuge at 3000r for 2min and remove the supernatant; add PBS to the rubber balls, shake and mix well, and then centrifuge at 3000r for 2min. This step is repeated ...
Embodiment 2
[0044] Example 2: Comparison of experimental group and control group in MCF-7 cells and MCF-10A cells, and analysis and comparison of 5-hmC content in MCF-7 cells and MCF-10A cells.
[0045] Add 50μl NE Buffer 4 to the lysed gel microspheres, shake and mix well, centrifuge at 3000r for 2min, remove the supernatant; add 2μM β-GT and 200μM UDP-N3-Glu to the gel beads, and carry out in 1×NE Buffer4 Reaction, 37°C, 500rpm shaking reaction for 3h; after the reaction, centrifuge at 3000r for 2min, remove the supernatant; add 50μl PBS to the glue ball, shake and mix well, then centrifuge, remove the supernatant, repeat this step three times; add to the glue ball 50μM DBCO-Cy5, in 1×PBS solution, 37°C, 500rpm shaking reaction for 2h in a dark environment; after the reaction, centrifuge at 3000r for 2min, remove the supernatant; add 50μl 2×SSC to the rubber ball, shake and mix evenly, and then centrifuge at 3000r for 2min, Remove the supernatant; add 20 μM SYBR-Green to the gel balls, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com