Primer probe group, kit and detection method for detecting fusobacterium nucleatum in saliva
A Fusobacterium nucleatum, primer probe technology, applied in the field of microbial molecular biology detection, can solve the problem of low accuracy of Fusobacterium nucleatum, achieve the effect of saving money, saving time, and improving detection efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0088]1. Extract the DNA in the saliva of 20 cases of healthy persons undergoing colonoscopy, patients with intestinal hyperplastic polyps, patients with adenoma, and patients with colorectal cancer. There was no significant difference in gender and age among the groups (P>0.05).
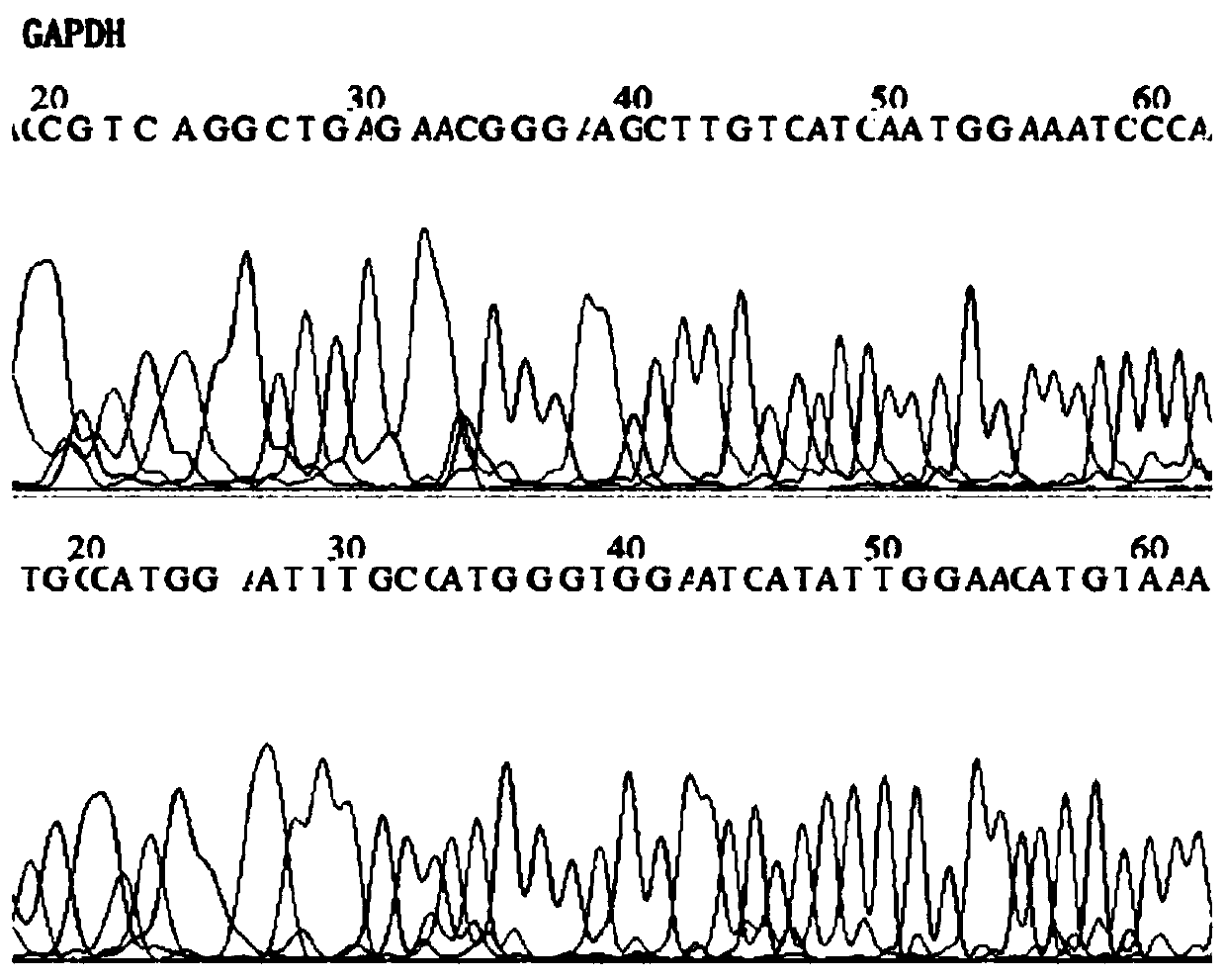
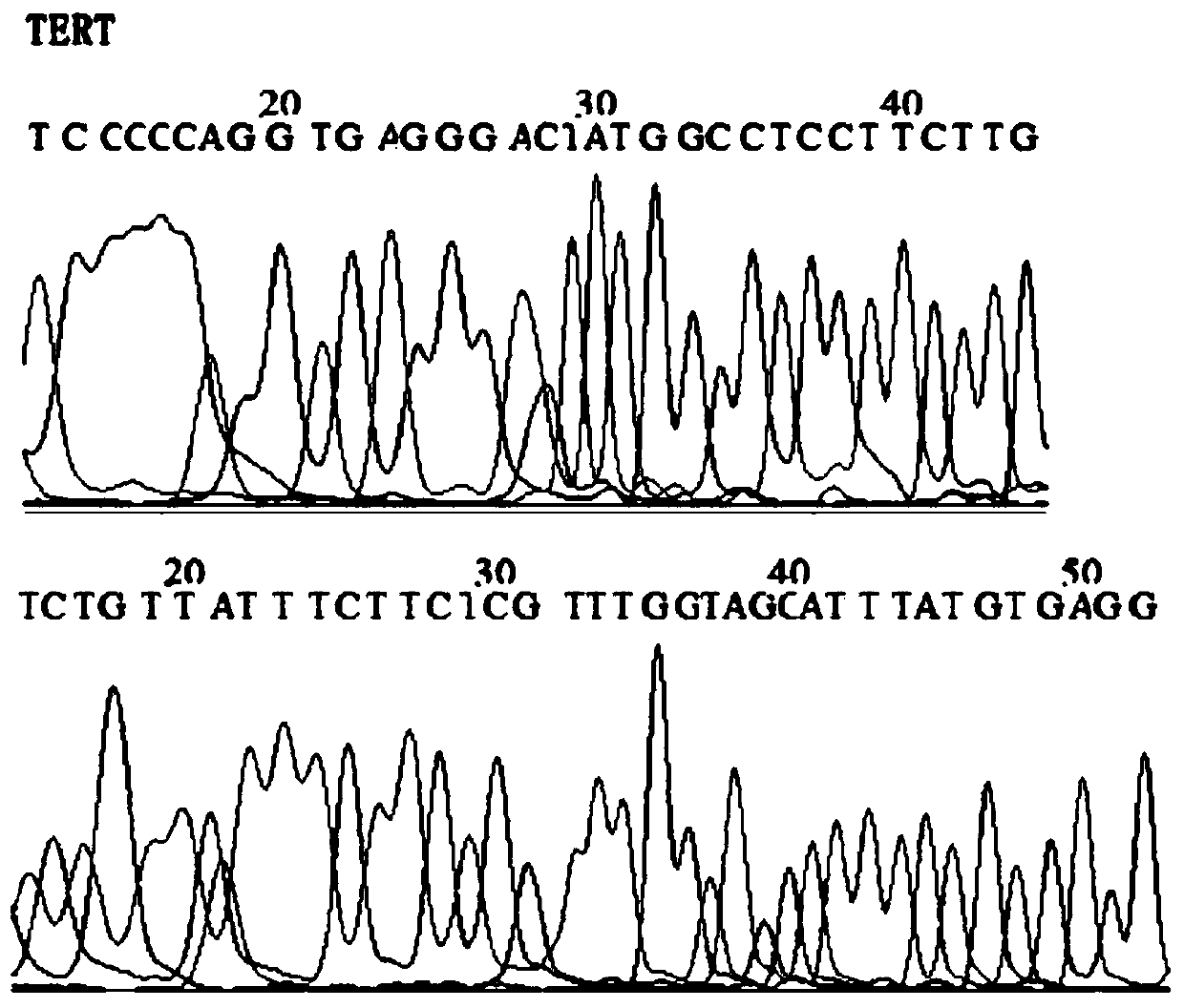
[0089] 2. 6 commonly used internal reference genes (GAPDH, RPPH1, NAGK, TERT, ERV-3, SLCO2A1) were detected by qPCR. After evaluation by NormFinder and geNorm software, it was found that the combination of GAPDH and TERT has a better effect on DNA quantification in saliva samples. Stability, see Table 1.
[0090] Table 1 Stability evaluation of internal reference genes
[0091]
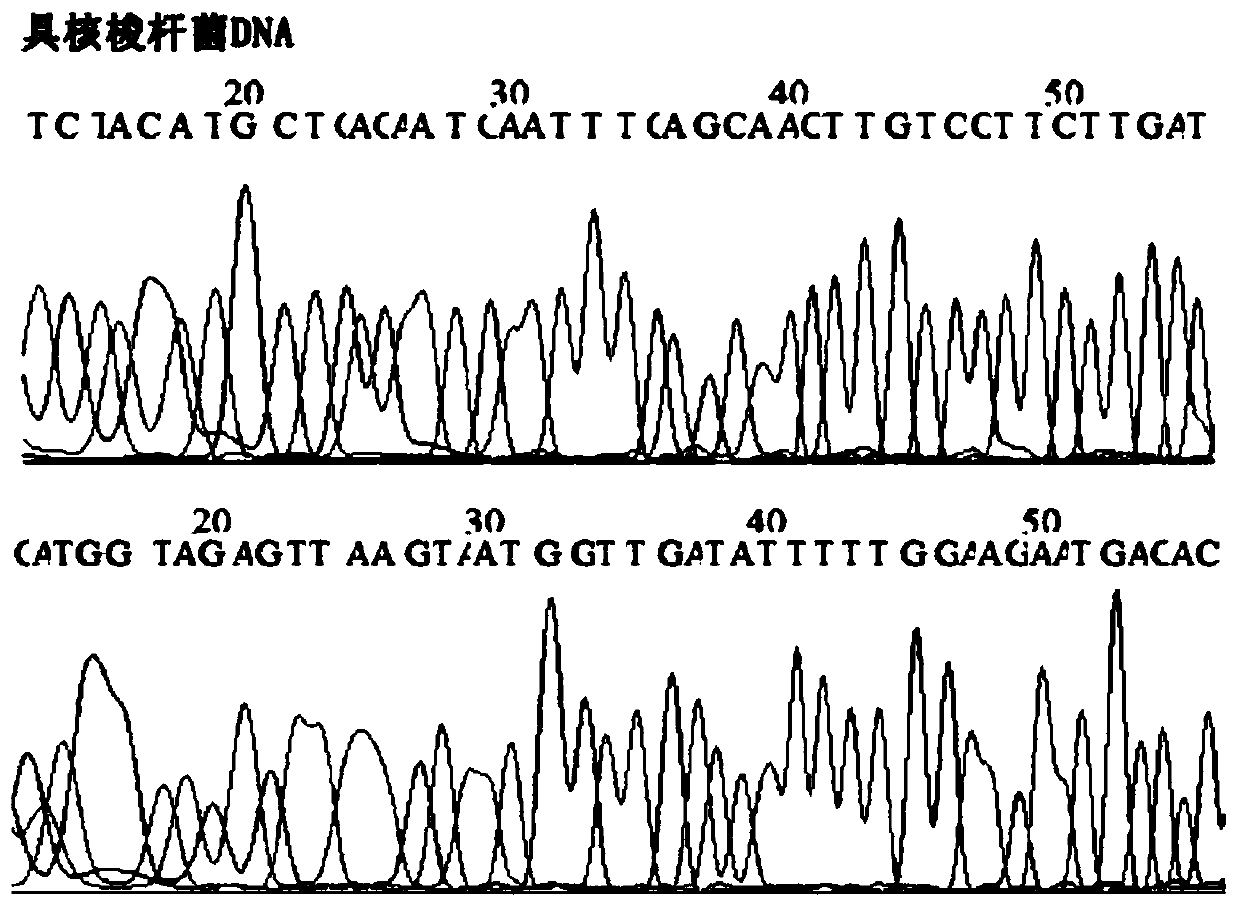
[0092] 3. Design primers and fluorescent probes for Fusobacterium nucleatum DNA and internal reference genes GAPDH and TERT. According to the TaqMan fluorescent probe PCR detection method, determine the design principles of primers and fluorescent probes to avoid mutual interference and interference between each primer ...
Embodiment 2
[0100] Case data: Saliva samples were collected from 20 healthy persons undergoing colonoscopy, patients with intestinal hyperplastic polyps, patients with adenoma, and patients with colorectal cancer. There was no significant difference in gender and age among the groups (P>0.05).
[0101] (3) Collection of serum samples: Use a saliva collection tube to collect the non-irritating whole saliva secreted by the patient's mouth after gargling with water, centrifuge at 3000rpm for 10min, then centrifuge at 11000rpm for 10-15min, collect the supernatant, and store it at -80°C.
[0102] (4) Extract DNA from saliva: Take about 200 μL saliva, add 20 μL proteinase K, 37 ° C for 1-2 hours; then add 200 μL buffer AL, vortex mix for 15 seconds, incubate at 56 °C for 10 min, add 200 μL absolute ethanol, vortex mix After standing still for 15s, vortex again for 15s, centrifuge, transfer the resulting mixture to a QLAamp micro spin column (in a 2mL collection tube), centrifuge at 6000g for 1...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com