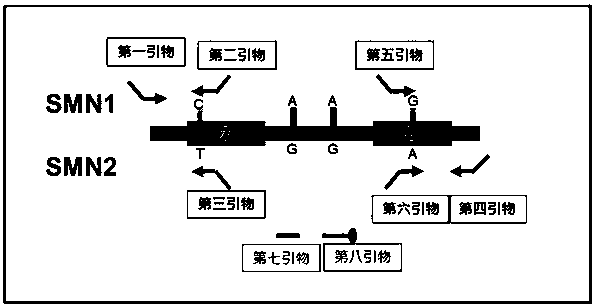
Detection kit for copy numbers of SMN1 and SMN2 genes
A technology for gene copy number and primer detection, which is applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc. The effect of improving detection specificity, simple operation and high detection sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] 1. Preparation of SMA reaction solution and SMA primer mixture
[0054] Table 1 SMA reaction solution component preparation list
[0055]
[0056] Table 2 SMA primer mixture component preparation table
[0057]
[0058] * ACTB upstream primer sequence:
[0059] TGACCGTCTGCGCCTCGTTCCATGTACGTTGCTATCCAGGC,
[0060] ACTB downstream primer sequence:
[0061] TCGACGCACGCTCCTGCTACAGCTCATTGCCAATGGTGATGAC,
[0062] HBB upstream primer sequence:
[0063] TGACCGTCTGCGCCTCGTTCACACAACTGTGTTCACTAGC,
[0064] HBB downstream primer sequence:
[0065] TCGACGCACGCTCCTGCTACATGGTCTCCTTAAACCTGTCTTG.
Embodiment 2
[0066] Embodiment 2 PCR amplification and result analysis
[0067] 1. Sample processing: Nucleic acid extractor (MagCore) and nucleic acid extraction kit (MagCore Genomic DNAWholeBlood Kit) extract human genomic DNA for subsequent PCR reaction, the DNA concentration is 2.5ng / uL~60ng / uL, the ratio of OD260nm / OD280nm Between 1.6-2.0.
[0068] 2. Preparation of amplification reagents:
[0069] (1) Take out the SMA reaction solution and SMA primer mixture from the kit, thaw at room temperature, mix them upside down, and centrifuge briefly with a microcentrifuge to make all the liquid settle to the bottom of the tube.
[0070] (2) Preparation of amplification reagents: prepare amplification reagents as shown in Table 3
[0071] Table 3 Amplification reagent preparation table
[0072]
[0073] * In order to reduce the separation error, it is recommended to take (n+1) parts of reaction solution and mixed enzyme according to the number of samples (n) when preparing amplification...
Embodiment 3
[0089] Example 3 Reagent Performance Verification
[0090] 1. Preparation of SMA normal control
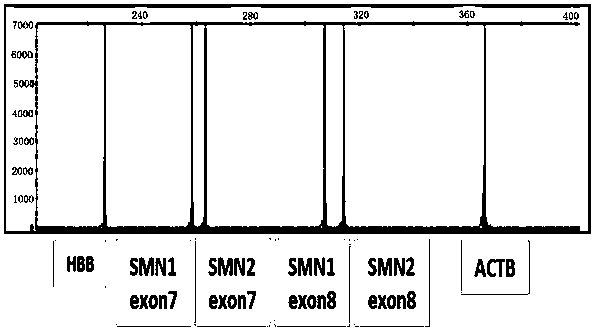
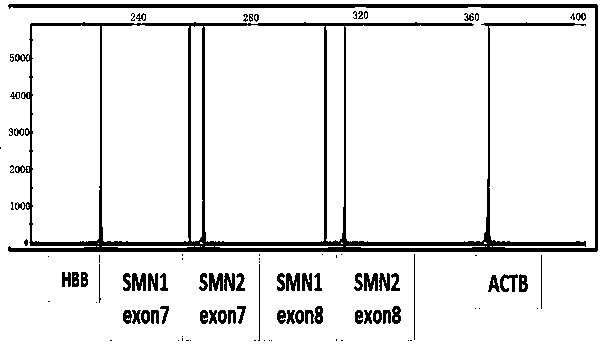
[0091] The 7th exon plasmid of SMN1 of 2 copy numbers, the 7th exon plasmid of SMN2 of 2 copy numbers, the 8th exon plasmid of SMN1 of 2 copy numbers, the 8th exon plasmid of SMN2 of 2 copy numbers, 2 The copy number of ACTB plasmid and the 2 copy number of HBB plasmid were respectively mixed in equal proportions, and the final concentration was 14500 copies / μL.
[0092] 2. Conformity rate of missing reference products
[0093] Detect 4 copies of missing reference products, as shown in the following table, respectively detect high, medium and low concentrations, the concentrations are 60 ng / uL, 25 ng / uL, 5 ng / uL respectively, and perform three repeated tests at each concentration, test three batches of reagents, and test results The positive coincidence rate was 100%.
[0094] Table 5 Gene copy numbers of exon 7 and exon 8 of deletion reference SMN1 and SMN2
[0095]
[009...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com