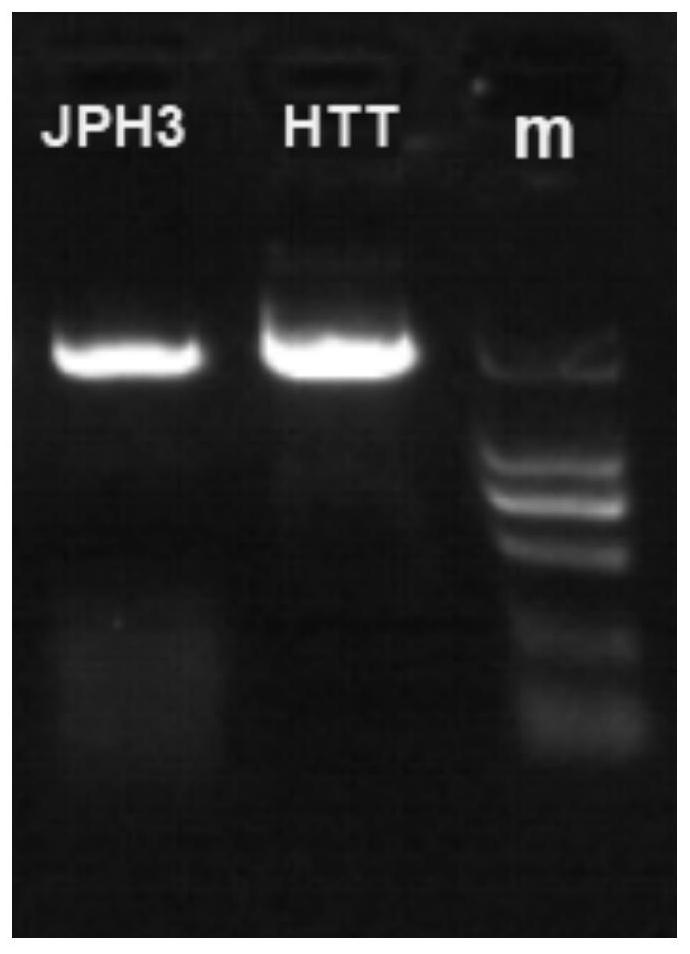
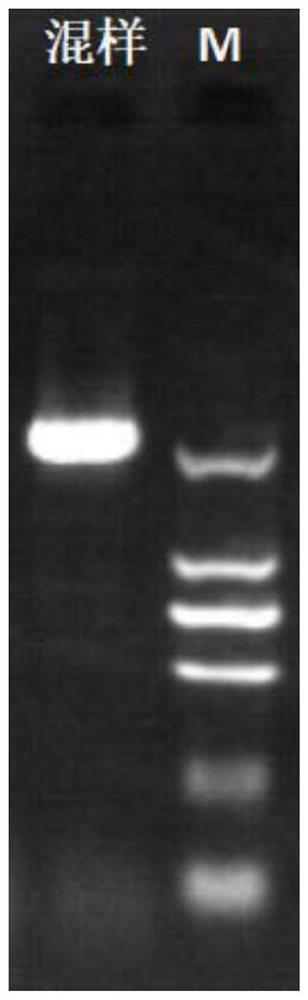
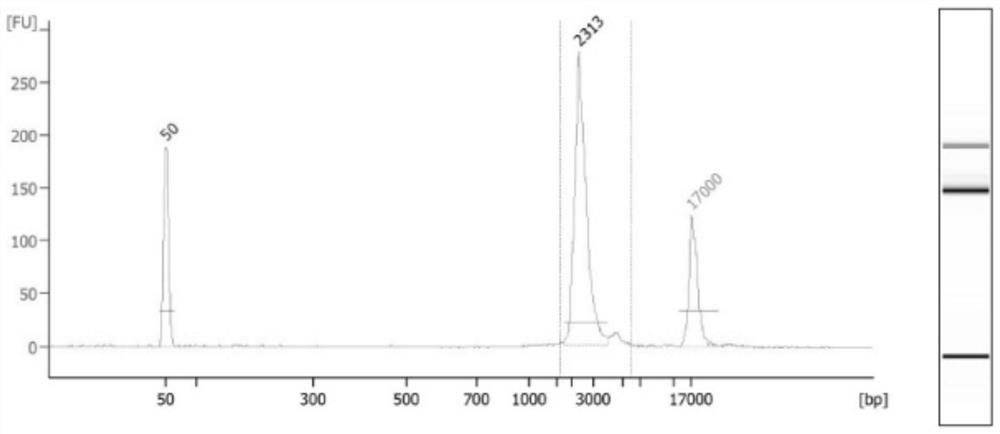
Detection primers, kits and methods for htt and jph3 genes
A kit and primer pair technology, applied in the field of biomedical detection and diagnosis, can solve the problems of CAG repeat sequence instability and incompletely clear mechanism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Example 1 DNA extraction and PCR amplification 1. DNA extraction experimental method
[0060] Use a kit to extract genomic DNA from blood, for example, use the TIANamp Blood DNA Kit Blood Genomic DNA Extraction Kit (DP348), and the operation is performed according to the kit instructions:
[0061] 1. Add Buffer CL to the blood sample, mix well and centrifuge;
[0062] 2. Discard the supernatant, add Buffer GS, mix well and centrifuge;
[0063] 3. Add Buffer GB and Proteinase K, mix well and incubate;
[0064] 4. Let stand at room temperature, add Buffer BD, and mix well;
[0065] 5. Pass through the adsorption column CG2, stand at room temperature, centrifuge, and discard the filtrate;
[0066] 6. Add Buffer BD to the adsorption column CG2, centrifuge, and discard the filtrate;
[0067] 7. Add Buffer PW to the adsorption column CG2, centrifuge, and discard the filtrate;
[0068] 8. Repeat step 7;
[0069] 9. Centrifuge and dry the adsorption column CG2;
[0070] ...
Embodiment 2
[0088] Example 2 Using PacBio sequal for third-generation sequencing
[0089] 1. Use the library building kit to build the library
[0090] 1 Repair the mixed library
[0091] Repair solution preparation:
[0092]
[0093] Mix well, centrifuge, put into PCR thermal cycler, and carry out repair reaction. The specific conditions are as follows:
[0094]
[0095]
[0096] 2 connector connection
[0097] The connection solution system is as follows:
[0098]
[0099] Mix well, centrifuge, and carry out the ligation reaction in a PCR thermal cycler. The specific conditions are as follows:
[0100]
[0101] 3 Purification
[0102] Use AMPure XP magnetic beads for purification, and finally use double-distilled water to elute the magnetic beads, and store at -20°C.
[0103] 2. Sequencing
[0104] 3. Bioinformatics analysis of sequencing data
[0105] 1.call ccs reads: apply the pacbio platform Smrtlink5-1 analysis process to call ccs reads.
[0106] 2. Barcode sp...
Embodiment 3
[0109] Example 3 Using the PromethION platform of Oxford Nanopore Technology for third-generation sequencing 1. Building a library using a library building kit
[0110] 1Library preparation
[0111] 1.1 End repair and A-tail ligation
[0112] Take the DNA, put it on ice, add NEB end repair and A-tail ligation reagents, and mix. Incubate for 40 minutes at 20°C and 20 minutes at 65°C.
[0113] 1.2 Magnetic bead purification
[0114] Add 1×AMPure beads to the DNA, incubate at room temperature for 15 minutes, adsorb on a magnetic stand at room temperature for 5 minutes, and discard the supernatant.
[0115] Add 80% ethanol, adsorb on a magnetic stand, discard the supernatant, and repeat once. Dry at room temperature.
[0116] Add Ultra Pure Water and elute by pipetting at 37°C.
[0117] Let stand on a magnetic stand for 5 min, and aspirate the supernatant, which is the purified DNA.
[0118] 1.3 Ligation of sequencing adapters
[0119] Add NEB T4DNA quick ligation buffer, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com