Antigenic polypeptide of poplar functional centromere histone cenh3 and its application
A technology of functional centromere and antigenic polypeptide, applied in the fields of bioinformatics and molecular cytogenetics, can solve the problem of increasing the difficulty of assembly of centromere sequences, and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0017] Example 1: Obtaining the amino acid sequence of the centromeric histone CENH3 of Populus trichocarpa
[0018] First, a gene Potri.014G096400.1 homologous to the gene encoding Arabidopsis CENH3 protein (At1g01370HTR12) was searched in the Populus trichocarpa genome database, that is, the CENH3 protein gene of Populus trichocarpa, thereby obtaining the amino acid sequence of the gene. The gene is located on chromosome 14 of P. trichocarpa and is a single copy in P. trichocarpa. Through the bioinformatics analysis of the amino acid sequence of CENH3 protein in different species, including Populus trichocarpa, Arabidopsis, rice and maize. The results showed that the amino acid sequence of Populus trichocarpa CENH3 protein was 54% similar to rice, 53% similar to maize, and 60% similar to Arabidopsis. The amino acid sequence of CENH3 proteins in different species varies greatly at the N-terminus but is relatively conserved at the C-terminus.
Embodiment 2
[0019] Example 2: Preparation of poplar centromere histone CENH3 antibody
[0020] The 20 specific amino acid sequences of the N-terminal of the centromere histone CENH3 protein of Populus trichocarpa were selected, the specific sequence was MARTKHPVARKRARSPKRSD, and the polypeptide was artificially synthesized according to the sequence as the antigen. The synthesized polypeptide was coupled to a protein carrier, and then routinely immunized New Zealand rabbits. After 4 immunizations, blood was collected from the rabbit carotid artery to collect serum. After the serum was passed through the antigen affinity column, rabbit-derived polyclonal antibodies were collected. The purity of the antibody was 90%, and the concentration of the antibody was 0.4mg / ml, the antibody was aliquoted and stored at -20°C.
Embodiment 3
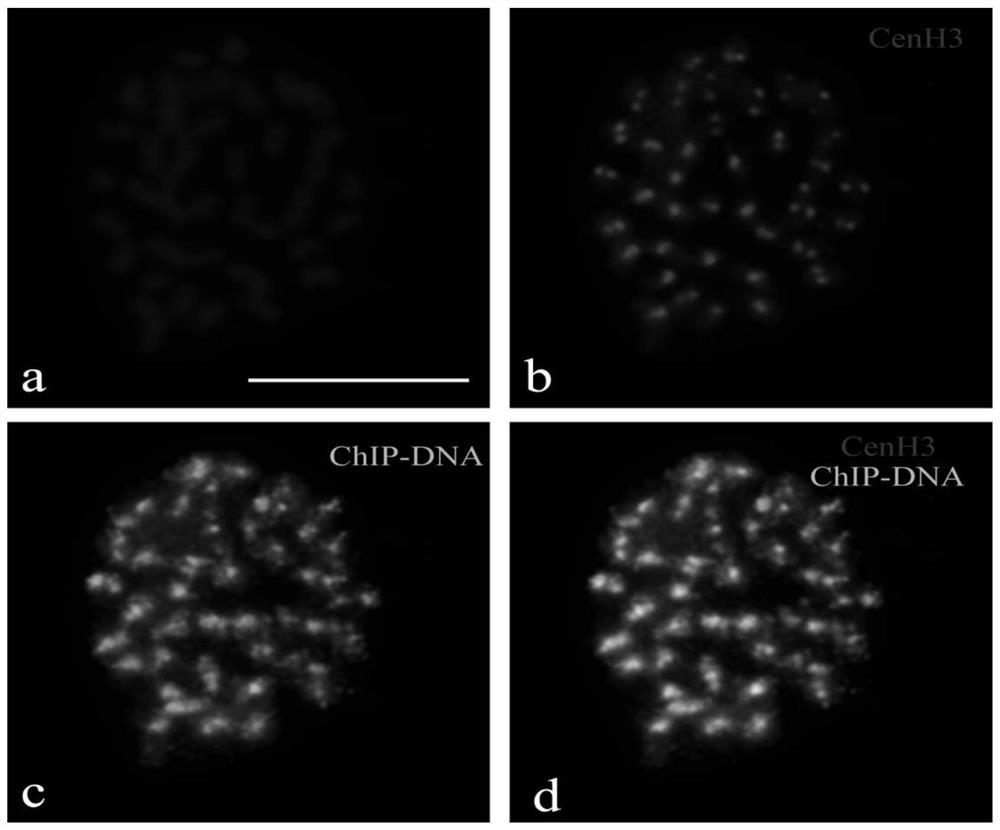
[0021] Example 3: Antibody Immunofluorescence Detection
[0022] Immunofluorescence detection is a cytological test for the specificity of the prepared antibody. Because the reaction between antigen and antibody is highly specific, the quality of the prepared CENH3 antibody can be judged by immunofluorescence detection. The experimental procedure is as follows:
[0023] (1) Root tip fixation: select vigorously growing root tips of poplar trees, fix with 4% (w / v) paraformaldehyde for 15 min, and then rinse with 1×PBS buffer solution for 3 times, 5 min each time.
[0024] (2) Chromosome preparation: 1) Transfer the root tip to a clean slide; 2) Cut off the root tip meristem with a double-sided blade; 3) Add 10 μL 1×PBS to the slide; 4) lightly Lightly cover the cover glass, tap the cover glass with a dissecting needle to fully disperse the apical meristem; 5) Soak the slide in a staining jar filled with 1×PBS, soak off the cover glass, take out the prepared The slices are air-...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com