Base editing molecule and applications thereof
A base editing and gene editing technology, applied in the field of fusion proteins
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
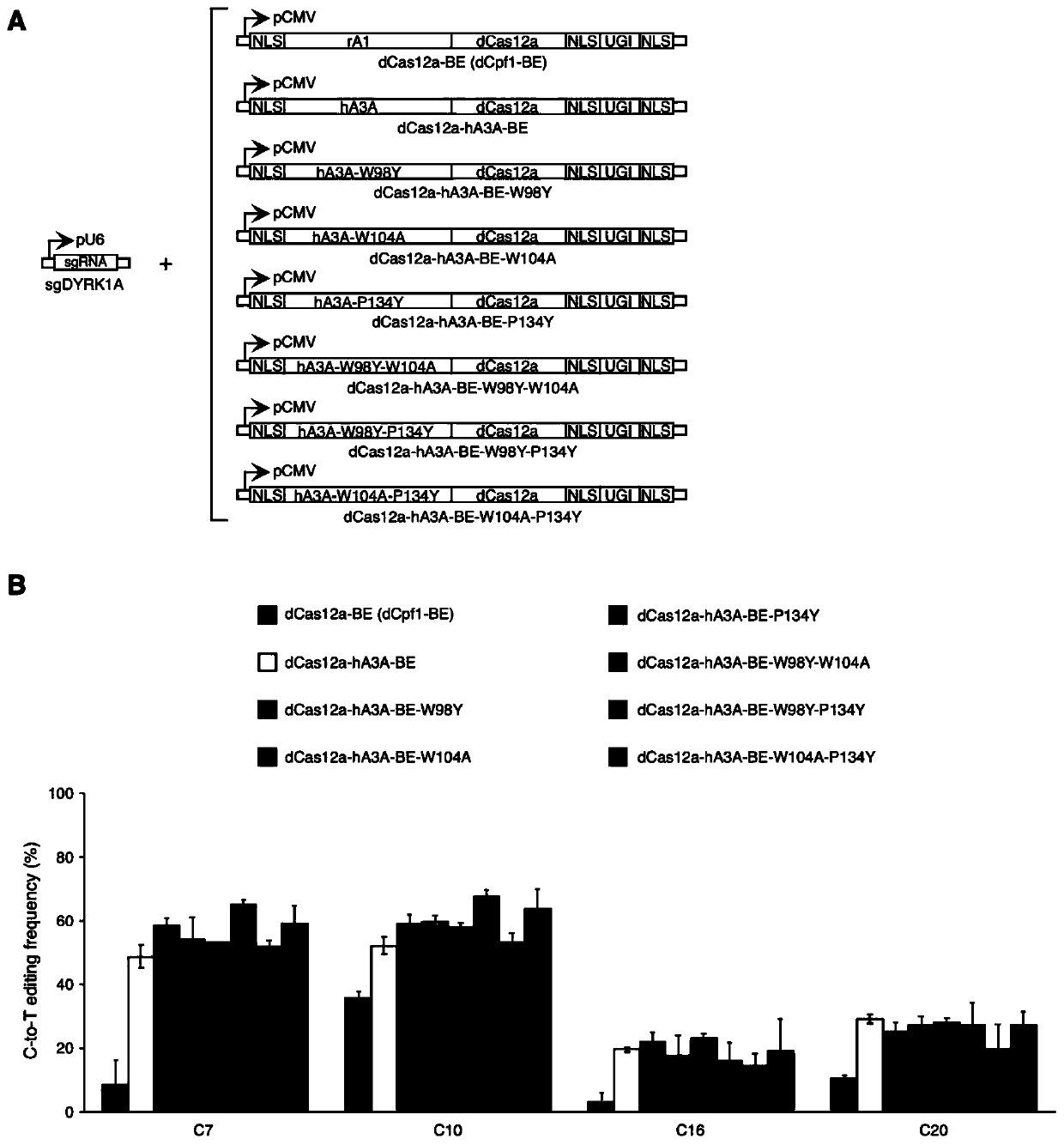
[0084] Example 1 Human genome DYRK1A target uses a series of dCas12a-hA3A base editors to achieve efficient and high-precision C-T base editing
[0085] 1.1 Experimental materials
[0086] Reagents and plasmids: primers synthesized from Sangon Bioengineering (Shanghai) Co., Ltd.; restriction enzymes, DNA ligase, high-fidelity DNA polymerase Purchased from NEB Company; Plasmid Recombination Kit Clone Purchased from Vazyme Company; DNA gel recovery kit was purchased from Corning; transfection reagent LTX, Available from Thermo Fisher; QuickExtract TM Genomic DNA extraction reagents were purchased from Illumina.
[0087] Cell line: Human fetal kidney cells HEK293T were cultured in DMEM medium (Gibco) supplemented with 10% fetal bovine serum (Gbico) and 1% double antibody (Gibco).
[0088] 1.2 Experimental method
[0089] Construction of dCas12a-hA3A-BE expression vector:
[0090] Using puc57-hA3A (synthesized by GenScript Biotechnology Co., Ltd.) as a template, using...
Embodiment 2
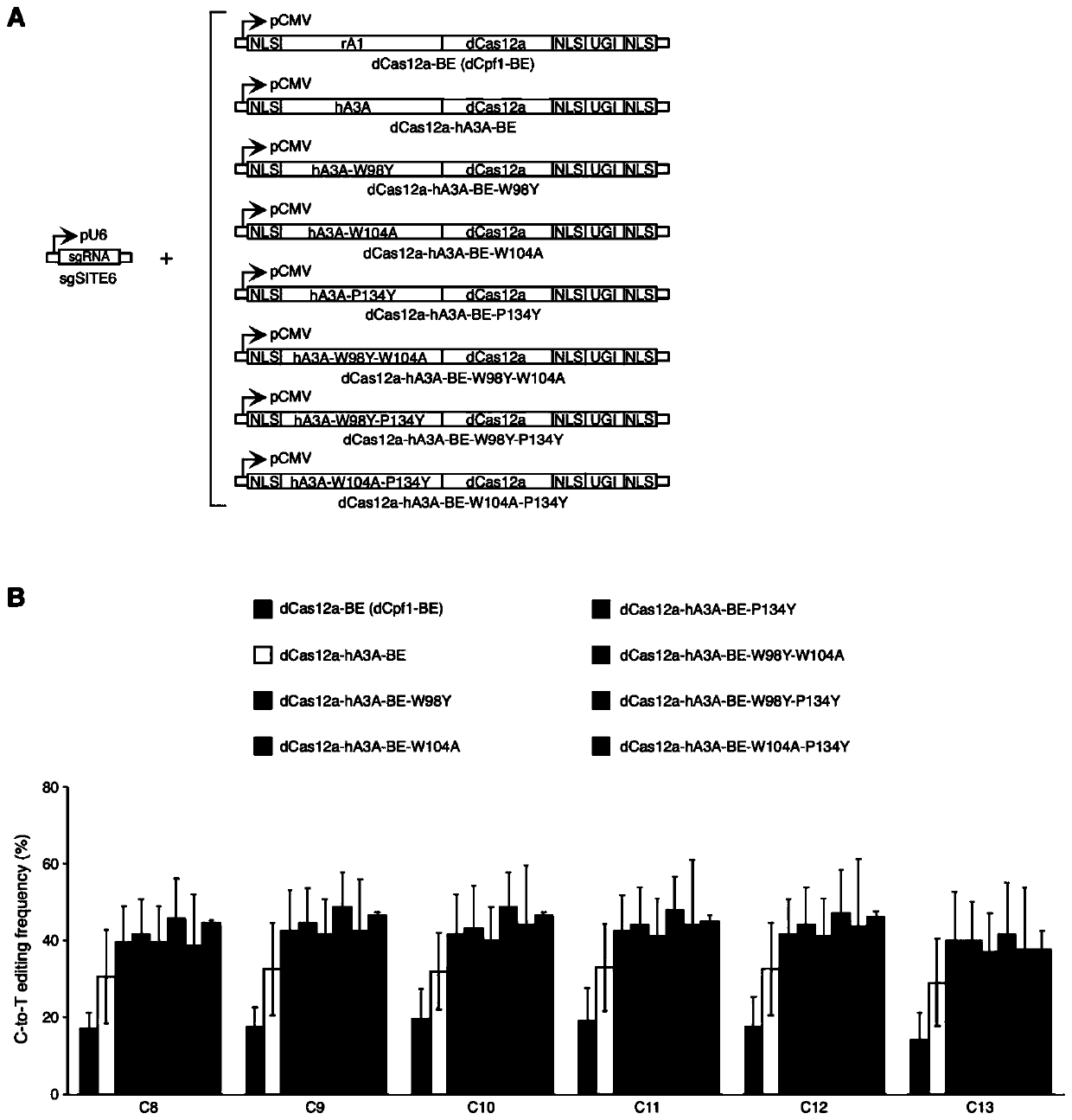
[0126] Example 2 The human genome SITE6 target uses a series of dCas12a-hA3A base editors to achieve efficient and high-precision C-T base editing
[0127] 2.1 Experimental materials
[0128] Reagents and plasmids: same as Example 1.
[0129] Cell line: same as Example 1.
[0130] 2.2 Experimental method
[0131] Construction of dCas12a-hA3A-BE expression vector: same as Example 1.
[0132] Construction of dCas12a-hA3A-BE-W98Y expression vector: same as Example 1.
[0133] Construction of dCas12a-hA3A-BE-W104A expression vector: same as Example 1.
[0134] Construction of dCas12a-hA3A-BE-P134Y expression vector: same as Example 1.
[0135] Construction of dCas12a-hA3A-BE-W98Y-W104A expression vector: same as Example 1.
[0136] Construction of dCas12a-hA3A-BE-W98Y-P134Y expression vector: same as Example 1.
[0137] Construction of dCas12a-hA3A-BE-W104A-P134Y expression vector: same as Example 1.
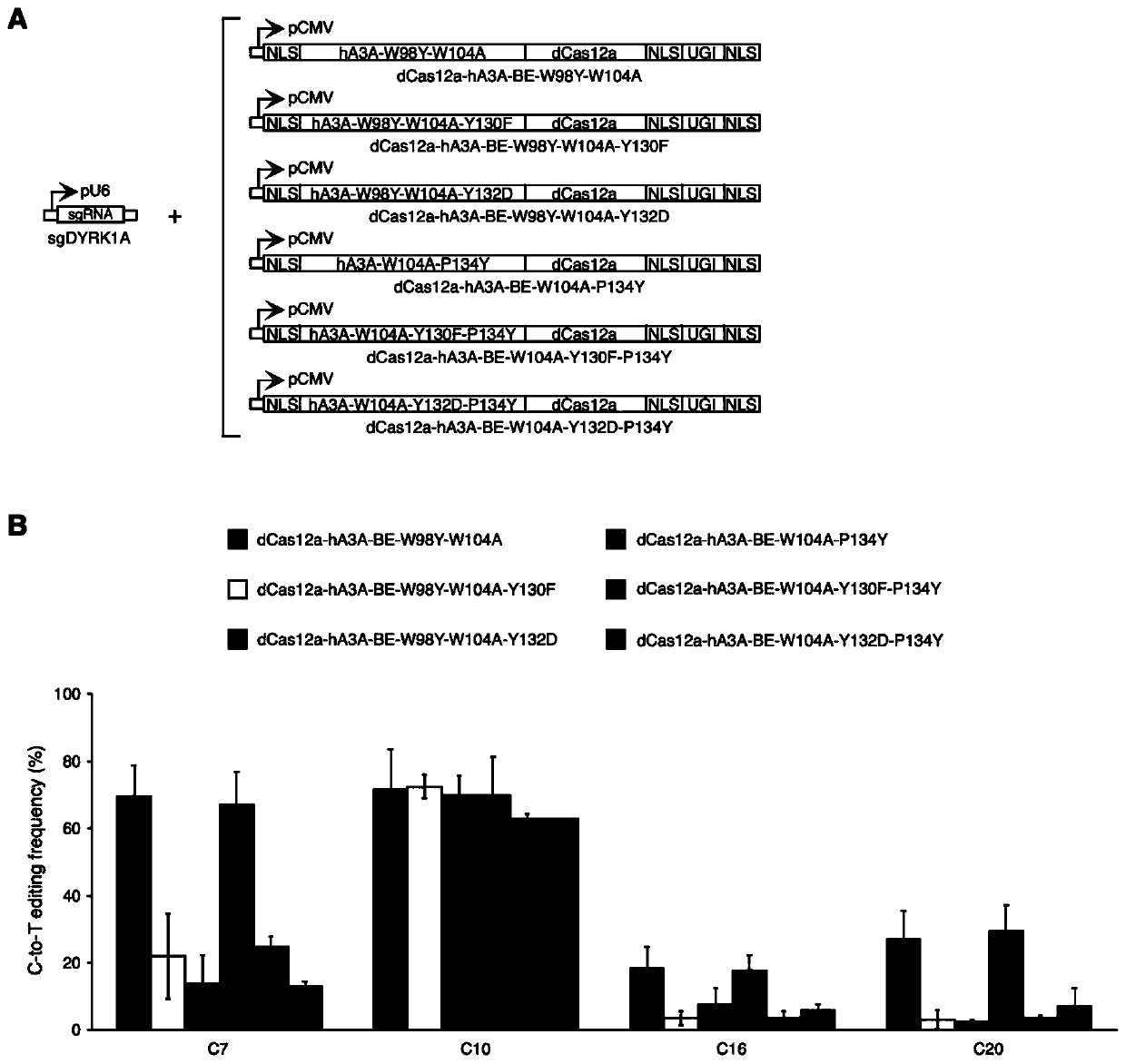
[0138] Construction of dCas12a-hA3A-BE-W98Y-W104A-Y130F expression vector...
Embodiment 3
[0156] Example 3 Human genome RUNX1 target uses a series of dCas12a-hA3A base editors to achieve efficient and high-precision C-T base editing
[0157] 3.1 Experimental materials
[0158] Reagents and plasmids: same as Example 1.
[0159] Cell line: same as Example 1.
[0160] 3.2 Experimental method
[0161] Construction of dCas12a-hA3A-BE expression vector: same as Example 1.
[0162] Construction of dCas12a-hA3A-BE-W98Y expression vector: same as Example 1.
[0163] Construction of dCas12a-hA3A-BE-W104A expression vector: same as Example 1.
[0164] Construction of dCas12a-hA3A-BE-P134Y expression vector: same as Example 1.
[0165] Construction of dCas12a-hA3A-BE-W98Y-W104A expression vector: same as Example 1.
[0166] Construction of dCas12a-hA3A-BE-W98Y-P134Y expression vector: same as Example 1.
[0167] Construction of dCas12a-hA3A-BE-W104A-P134Y expression vector: same as Example 1.
[0168] Construction of dCas12a-hA3A-BE-W98Y-W104A-Y130F expression vector: ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com