RNA fixed-point editing by artificially constructing RNA editing enzyme and related application
An editing enzyme and editing technology, applied in the field of RNA-directed editing and related applications, can solve the problems of difficult transfer of genomic loci, low-efficiency gene introduction, and long-term safety concerns of genomic DNA editing.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
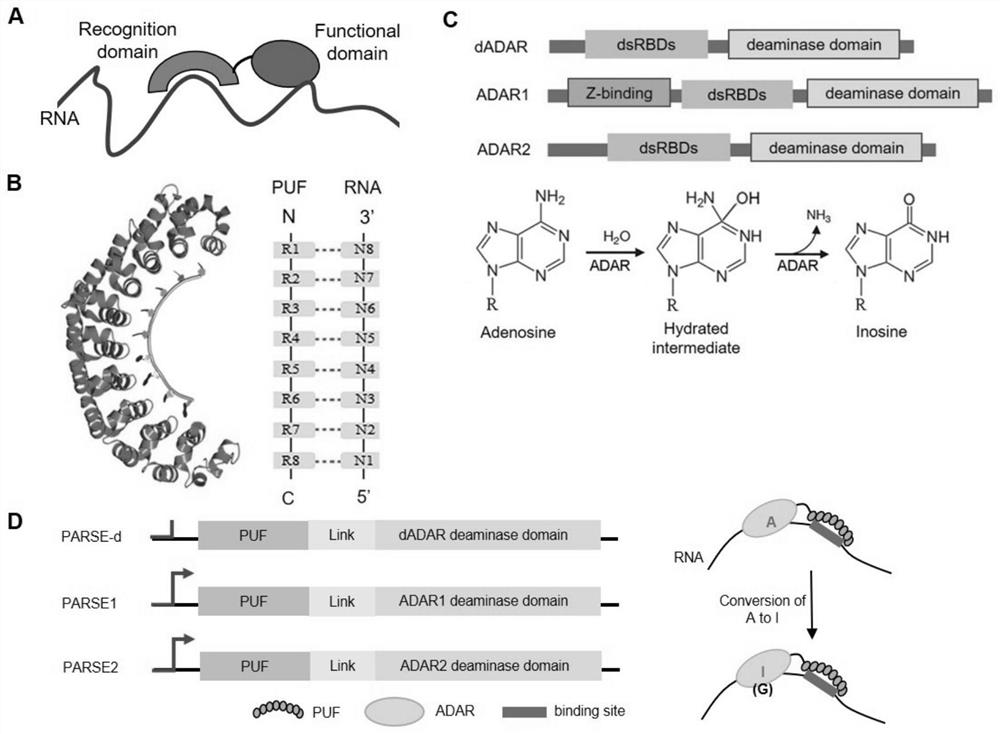
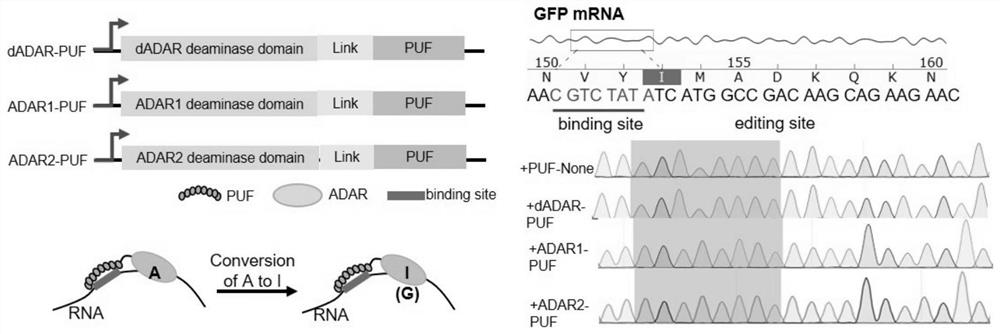
[0226] ADAR is an adenosine editing enzyme that can catalyze the deamination of RNA adenosine to form inosine (adenosine-to-inosine, A-to-I). Three ADAR catalytic domains from different sources were cloned, and the linker and RNA binding protein PUF fusion, developed three different artificial RNA editing enzymes (RNA editase), named the system as artificial P UF- A DAR R NA s sequence e ditors(PARSE-d,PARSE1,PARSE2)( figure 1 )
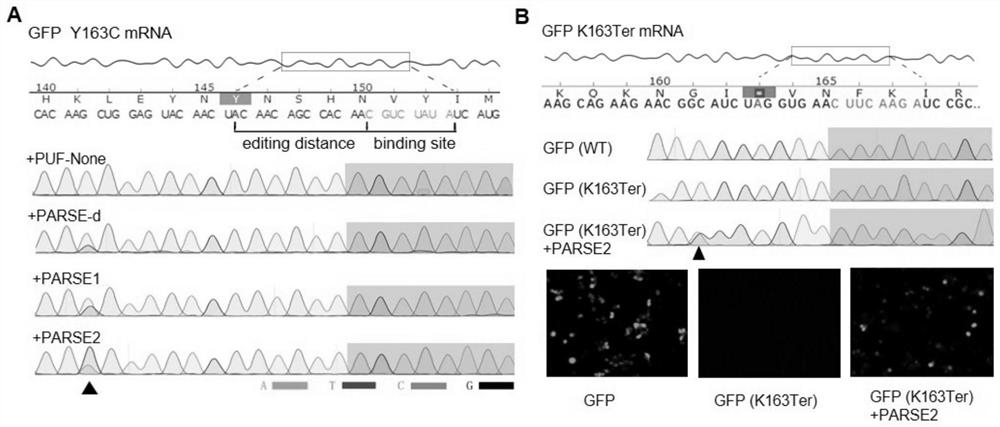
[0227] Detect the editing activity of the three RNA editing enzymes PARSE-d, PARSE1 and PARSE2 at the cellular level, transfer the PARSE RNA editing enzyme and the GFP reporter gene plasmid into the cells at the same time, collect the RNA of the GFP reporter gene after 48 hours and sequence it, and detect RNA editing events , proving that PARSE-d, PARSE1, and PARSE2 all have the ability to efficiently edit target RNA ( figure 2 A). By constructing a GFP mRNA containing an early stop codon, PARSE is used to realize the editing of A to G at a s...
Embodiment 2
[0230] The editing efficiency of ADAR on target RNA is improved by optimizing the ADAR catalytic domain, and the efficiency, precision and off-target rate of RNA editing enzymes are detected and analyzed by RNA seq high-throughput sequencing technology. A new point mutation was introduced in each of the catalytic domains of ADAR1 and ADAR2 to improve the editing efficiency of ADAR ( Figure 4 A), using RNA seq high-throughput sequencing technology to detect and analyze the efficiency and accuracy of RNA editing enzymes, and found that PARSE1, ePARSE1, PARSE2, and ePARSE2 can all edit target RNA, with efficiencies of 42%, 65%, and 67% respectively , 78% ( Figure 4 B). The analysis of RNA-seq results shows that there is a certain degree of off-target rate in PARSE editing, but compared with the editing efficiency of the target site, the off-target efficiency is lower, and the off-target rate can be reduced by reducing the transfection amount of PARSE in the later optimization ...
Embodiment 3
[0234] Optimize the RNA-binding protein PUF to improve the precision of PARSE editing RNA and reduce off-target efficiency. By optimizing PUF, PUF8, which recognizes 8 bases, is optimized to PUF10, which recognizes and binds to ten bases ( Figure 7 ), did not reduce the editing efficiency at the target site, but as the number of PUF recognition and binding bases increased, we believe that this strategy can effectively reduce off-target effects, and has the potential for further optimization, we can optimize PUF to recognition And combining 12 bases, 16 bases, or even longer, can greatly expand the scope of application of PARSE.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com