CircPTEN1 for tumor treatment target and diagnosis biomarker and application of circPTEN1
A biomarker, tumor treatment technology, applied in circPTEN1 and its application fields, can solve the problem of unclear specific role
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] Discovery and identification of circular RNA (circPTEN1) derived from PTEN gene
[0055] In this implementation case, the existence of circular RNA (circPTEN) derived from the PTEN gene was determined by using biological information means combined with PCR amplification technology and its biological characteristics were identified. The specific content is as follows:
[0056] 1. Discovery of circular RNA (circPTEN) derived from PTEN gene
[0057] Using the circBase (http: / / www.circbase.org) database analysis, it was found that the PTEN gene can be spliced to form 8 kinds of circRNAs, namely hsa_circ_0002232, hsa_circ_0002934, hsa_circ_0003058, hsa_circ_0019058, hsa_circ_0019059, hsa_circ_0019060, hsa_4circ_3 and 0hsa_4circ_4.
[0058] Different from the traditional linear splicing of PTEN mRNA, hsa_circ_0002232 is encoded by the PTEN gene (chr10:89624210-89693008) The second half of the first exon and the 2nd, 3rd, 4th, 5th exons through reverse spliced, and its matu...
Embodiment 2
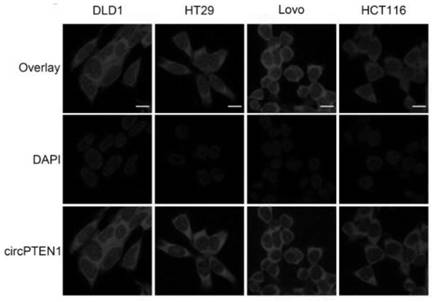
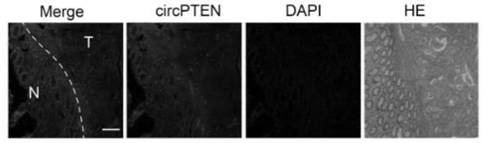
[0094] Expression of circPTEN in colorectal cancer tissues
[0095] In this implementation case, the use of qRT-PCR and FISH experiments revealed that the expression of circPTEN in colorectal cancer tissues was down-regulated and related to the clinicopathological indicators and prognosis of patients. The details are as follows:
[0096] 1. qRT-PCR and FISH experiments reveal that circPTEN is down-regulated in colorectal cancer tissues
[0097] 1.1 qRT-PCR revealed that the expression of circPTEN was decreased in colorectal cancer tissues. The previous identification experiments of circPTEN revealed that circPTEN is an abundant and stable circRNA expressed in colorectal cancer produced by PTEN through back splicing. In order to further explore the relationship between circPTEN and colorectal cancer, the expression level of circPTEN was detected by qRT-PCR in 150 pairs of fresh colorectal cancer tissues and their paired paracancerous tissues (the primer sequences were SEQ ID N...
Embodiment 3
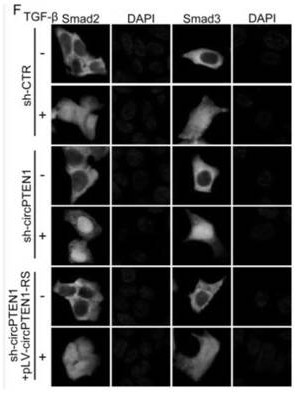
[0109] In this implementation case, experiments such as RNApulldown, RIP, Western blot, and qRT-PCR revealed that the expression level of circPTEN1 is regulated by EIF4A3, and the details are as follows:
[0110] In order to explore why the expression level of circPTEN1 is down-regulated in colorectal cancer tissues, that is, to explore the specific factors that regulate the expression level of circPTEN1, we first analyzed the formation mechanism of circRNA. The formation mechanism of circRNA mainly includes intron pairing-driven circularization and RBP-driven circularization, that is, the upstream and downstream flanking introns of circRNA contain Alu sequences with reverse complementary sequences or RBP is combined in the flanking introns, both It can regulate the splicing and circularization of circRNA. Many RBPs (such as QKI, FUS, and ADAR1, etc.) can bind to the intron sequences around the circular exon, thereby promoting or inhibiting the circularization of circRNA. How...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com