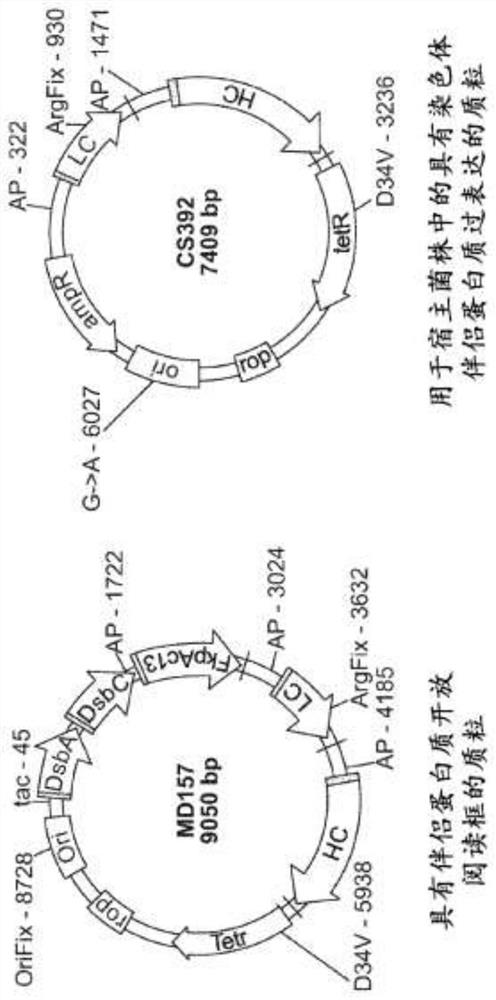
Methods of producing two chain proteins in prokaryotic host cells
A host cell, protein technology, applied in the field of prokaryotic host cells, can solve the problem of increasing complexity of recombinant therapy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0297] The present disclosure will be understood in more detail with reference to the following examples. However, they should not be construed as limiting the scope of the present disclosure. It should be understood that the examples and embodiments described herein are for illustrative purposes only, and that various modifications or changes thereof will be suggested to those skilled in the art, and will be included within the spirit and scope of this application and the scope of the appended claims. within range.
example 1
[0298] Example 1: Engineering E. coli strains with chromosomally integrated promoters controlling the expression of the chaperone proteins DsbA, DsbC and FkpA
[0299] Overexpression of the chaperone proteins DsbA, DsbC and FkpA from plasmids improves the production of antibody-based products in bacterial culture. However, expression of these chaperone proteins from plasmids has several disadvantages. For example, this approach requires the development and regulation of expression plasmids for each new product. In some cases, large plasmid sizes can also lead to lower product titers. Additionally, plasmids are typically present in 10-15 copies per cell, resulting in high levels of overexpression, which may require downstream purification steps to remove chaperone proteins (eg, FkpA) from the product. In some cases, lower expression levels of one or more chaperone proteins can achieve the same product titers.
[0300] Here, the native promoters of dsbA, dsbC, and fkpA in the...
example 2
[0321] Example 2: Overexpression of FkpA under the control of a chromosomally integrated promoter
[0322] Overexpression of FkpA was compared in strains engineered as described in Example 1 with an integrated promoter overexpressing native FkpA compared to strains carrying a plasmid expressing FkpA.
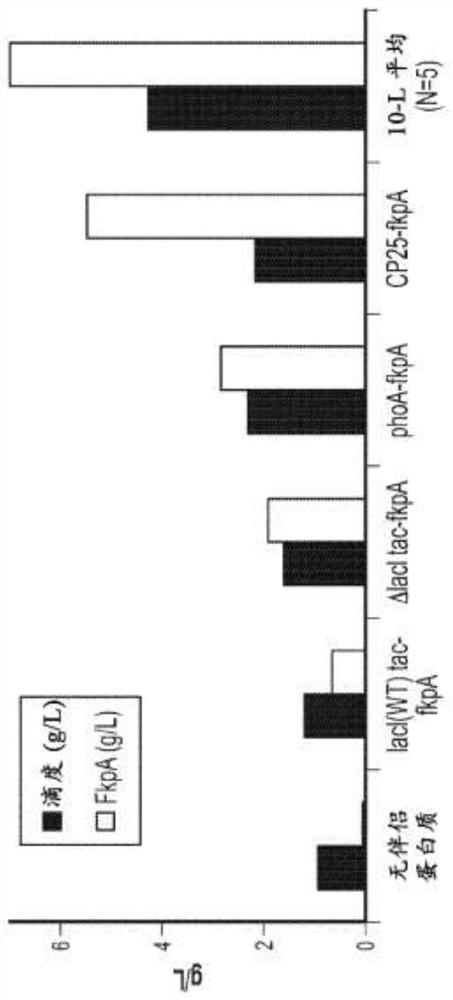
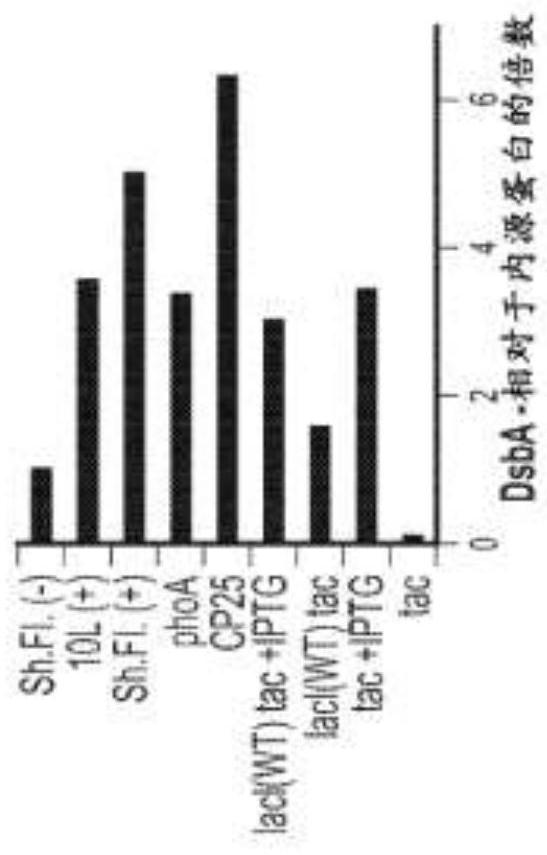
[0323] To assess the ability of FkpA to be overexpressed in the chromosome, the native promoter of FkpA was exchanged with the tac, phoA or CP25 promoter according to the method of Example 1. The lacI promoter was also modified to further enhance the strength of the tac promoter. Strains were grown in 10 L fermentations and FkpA production was assessed by Western blotting.
[0324] Chromosomal expression driven by the CP25 promoter showed the highest level of FkpA expression, followed by expression driven by the phoA promoter ( figure 2 ). As expected, the tac promoter showed the lowest level of FkpA expression, with higher FkpA expression in the ΔlacI background than in the...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com