Plasmid constructs for treating cancer and methods of use
A technology of plasmids and structural domains, applied in chemical instruments and methods, treatment, electrotherapy, etc., can solve the problems of increasing the existence and quantity of tumor-specific T cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0253] CXCL9
example 1
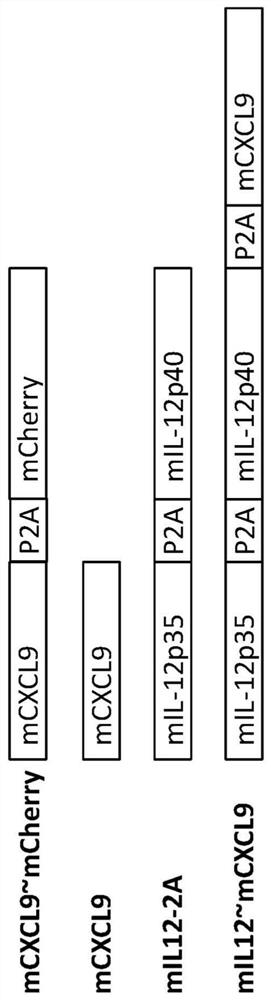
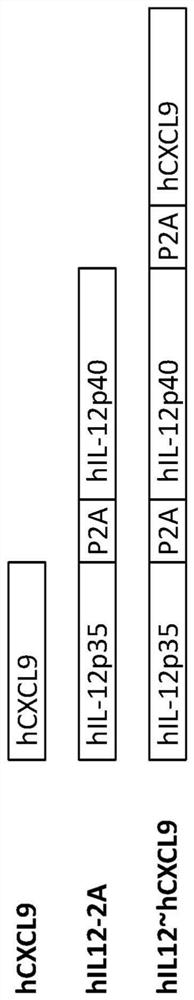
[0254] Example 1. CXCL9 plasmid construction. Mouse CXCL9 (mCXCL9) or human CXCL9 (hCXCL9) nucleic acid sequences were cloned into expression vectors using standard molecular biology techniques. Alternatively, mCXCL9 or hCXCL9 was cloned downstream of mouse (mIL12-2A) or human (hIL12-2A) IL12 p35-P2A-IL12 p40 to generate mIL12~mCXCL9 and hIL12~hCXCL9 ( Figure 1A -B). The IL12 p35-P2A-IL12 p40 construct was prepared essentially as described in WO2017 / 106795 or WO2018 / 229696.
[0255] The resulting plasmid contained IL-12p35, IL-12p40, and CXCL9, all expressed from the same promoter, with an intermediate exon-skipping (P2A) motif, allowing all three proteins to be expressed from a single polycistronic information expression. Using a similar method to prepare mCXCL9~mCherry
example 2
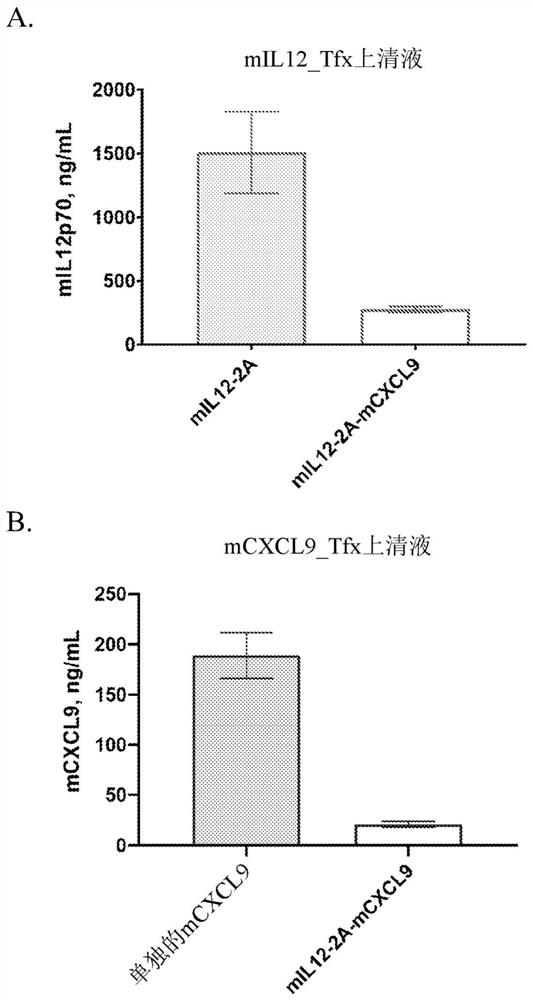
[0256] Example 2. Protein expression. The mIL12-2A, mCXCL9 and mIL12-mCXCL9 expression vectors were transfected into HEK293 cells in vitro. 96 hours after transfection, supernatants were collected and assayed for IL12 and CXCL9 protein expression by ELISA. figure 2 The results shown in , show that despite decreased expression in cells transfected with the mIL12~mCXCL9 expression vector, detectable levels of both IL12 and CXCL9 were produced. Figure 27 High levels of secreted hIL12 and hCXCL9 were shown in cells transfected with hIL-12~hCXCL9 expression vectors.
[0257] Similarly, hIL12-2A, hCXCL9 and hIL12~hCXCL9 expression vectors were transfected into HEK293 cells in vitro. 96 hours after transfection, supernatants were collected and assayed for IL12 and CXCL9 protein expression by ELISA. hIL12 was almost equally expressed by both hIL12-2A (1.59 μg / mL) and hIL12~hCXCL9 (1.37 μg / mL) expression vectors ( Figure 10 A). Compared with cells transfected with hCXCL9 expres...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com