DNA methylation biomarkers for early detection of cervical cancer
A biomarker and early detection technology, applied in the field of molecular diagnosis, can solve problems such as hindering disease manifestations, and achieve the effect of specific and sensitive risk assessment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example
[0208] The following examples are given for the purpose of illustrating the invention and therefore should not be construed as limiting the scope of the invention.
example 1
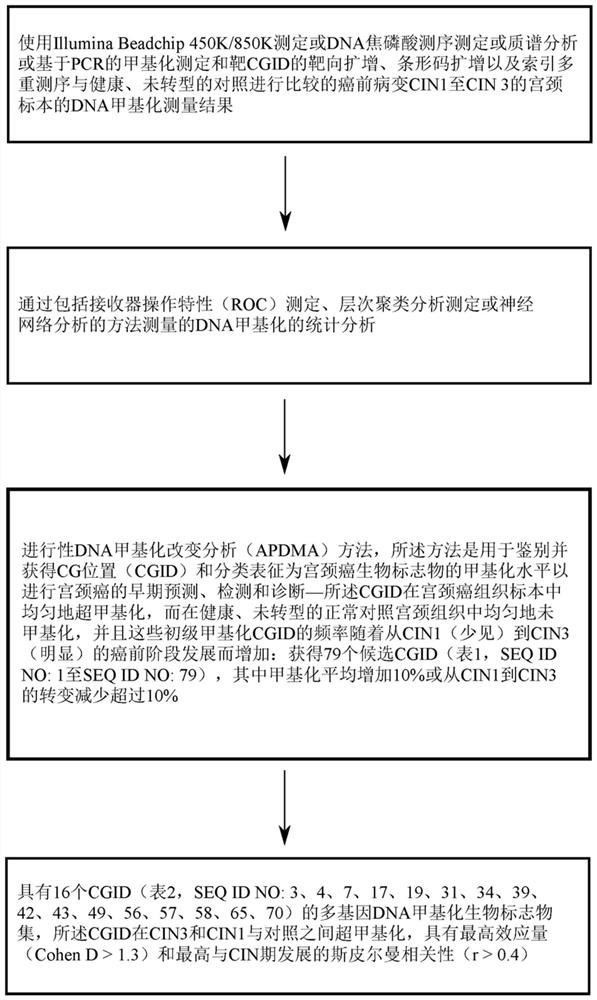
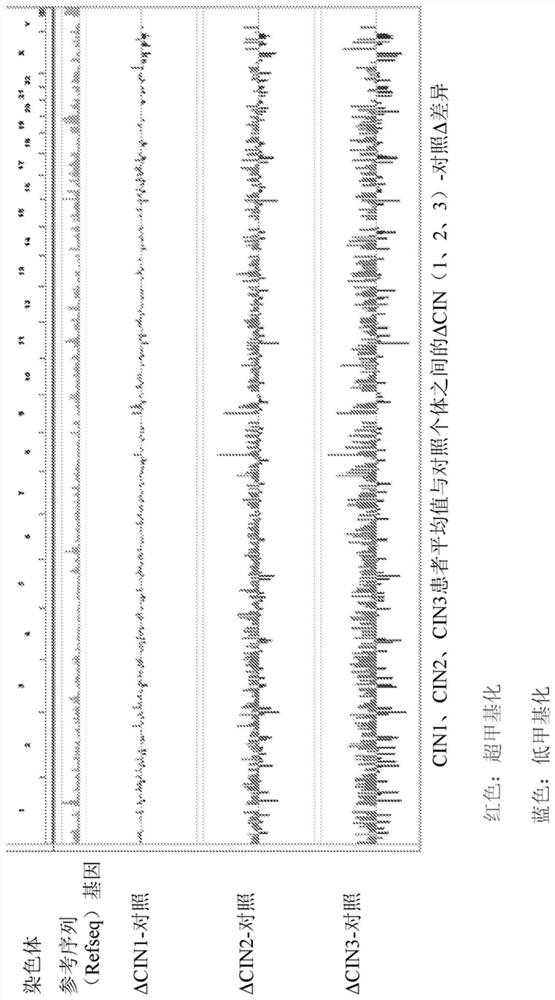
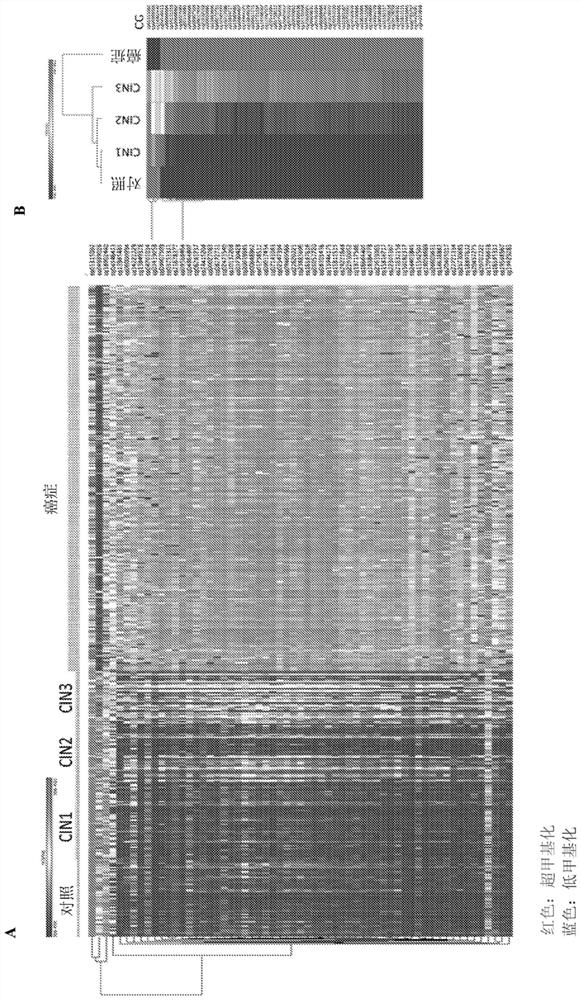
[0209] Example 1: Progressive DNA methylation alteration analysis (APDMA) method for determining and obtaining CG position (CGID), the methylation level of CG position is an early predictor of cervical cancer.
[0210] The present invention addresses a major challenge in cervical cancer screening, namely finding robust biomarkers that provide highly accurate and sensitive risk assessments that can guide early intervention and treatment. A general approach is to use case-control logistic regression on genome-wide DNA methylation data to identify loci that are more or less methylated in cancer cells than in controls. However, it is well known that many of the cancers detected by these methods with statistically significant DNA methylation changes are heterogeneous, and many occur late in the development of the cancer and are therefore of very limited value in early detection, which is because they are diluted when the frequency of cancer cells in the specimen is low. Furthermor...
example 2
[0225] Example 2: Discovery of a multigene DNA methylation biomarker set for early detection of cervical cancer.
[0226] The present disclosure further selected 16 CGIDs from the list obtained and disclosed in the first example and Table 1, wherein the 16 CGIDs were hypermethylated between CIN3 and CIN1 and the control, and between CIN3 and the control The highest effect between patients (Cohen D>1.3) and the highest Spearman correlation with CIN stage development r>0.4. (See Table 2 above).
[0227] Next, to obtain the minimum number of CGIDs required to distinguish CIN3 precancerous lesions from control lesions, our method performed penalized regression to reduce the number of CGIDs to five. Then, the method performed multivariate linear regression with these 5 CGIDs as independent variables and CIN3 status as dependent variable. Both CGIDs remained significant (see Table 3 above). A linear regression equation consisting of the weighted methylation levels of these two lo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com