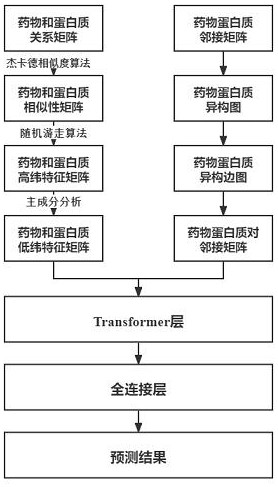
Graph-Transform-based drug target interaction prediction method research
A prediction method and drug technology, applied in the field of biochemistry, can solve the problems of not considering the graph Transformer, not applying the graph structure, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
example 1
[0030] Example 1 The present invention is tested by the long text news collected by oneself
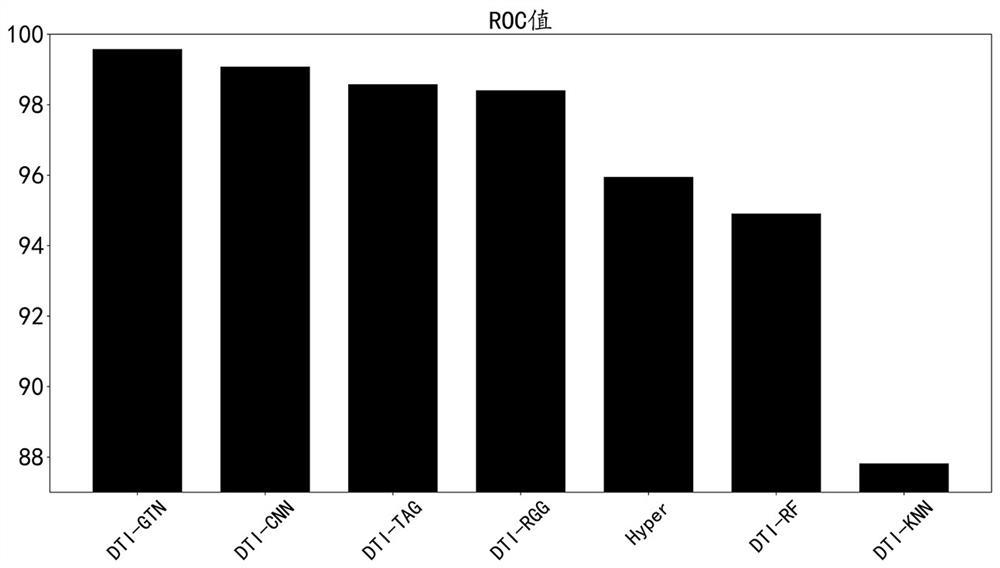
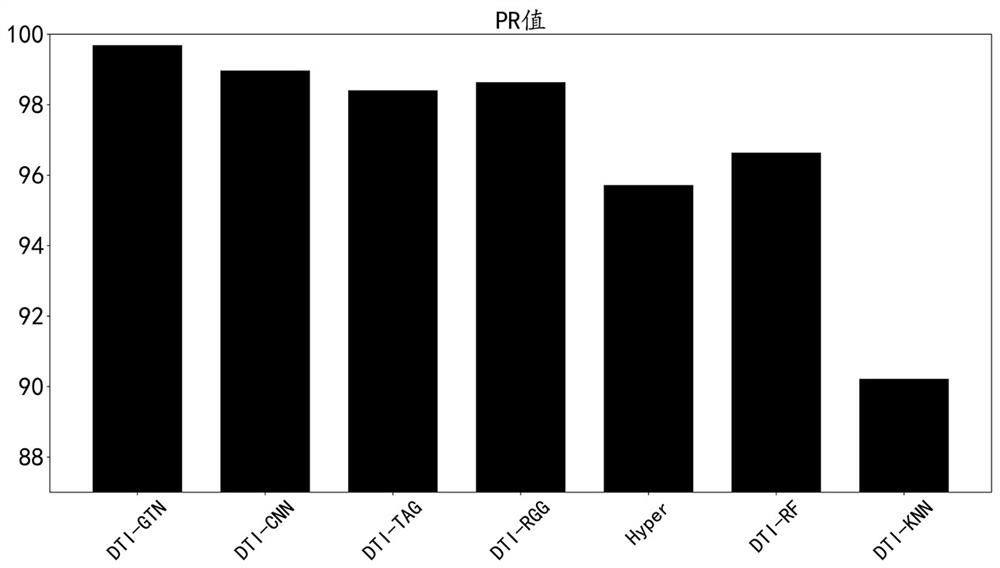
[0031] This dataset is a dataset consisting of 12,015 nodes and 1,895,445 edges, and is a dataset for drug-target interaction prediction tasks. The dataset integrates four types of nodes (drug, protein, disease, and side effect) and five types of edges (drug-drug interaction, drug-disease association, drug-side effect association, protein-disease association, and protein-protein interaction). The present invention selects the DTI-GTN model as the basic model of the drug target prediction model, and uses two indicators to evaluate its performance, namely Receiver Operating Characteristic Curve (ROC) and Precision-Recall (PR). For comparison, they are DTI-GTN, DTI-CNN, DTI-TAG, DTI-RGG, DTI-Hyper, DTI-RF, DTI-KNN. The existing 6 methods all run under their optimal parameters. The relevant parameters of the method of the present invention are set as follows: the number of epochs is 20...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com